Abstract
BACKGROUND AND OBJECTIVES:
A new test (Dr. KSU H1N1 RT-PCR kit) was recently developed to provide a less expensive alternative to real-time reverse transcriptase-polymerase chain reaction (RT-PCR). We report the findings of a validation study designed to assess the diagnostic accuracy, including sensitivity and specificity, of the new kit, as compared to real-time RT-PCR.
DESIGN AND SETTING:
Cross-sectional validation study conducted from 18-22 November 2009 at a primary care clinic for H1N1 at a tertiary care teaching hospital in Riyadh.
PATIENTS AND METHODS:
Nasopharyngeal swab samples and data on socio-demographic characteristics and symptoms were collected from 186 patients. Swab samples were sent to the laboratory for testing with both real-time RT-PCR and the new Dr. KSU H1N1 RT-PCR kit. We measured the sensitivity and specificity of the new test across the entire sample size and investigated how these values were affected by patient socio-demographic characteristics and symptoms.
RESULTS:
The outcomes of the two tests were highly correlated (kappa=0.85; P<.0001). The sensitivity and specificity of the new test were 99.11% and 83.78%, respectively. The sensitivity of the new test was affected only minimally (96%-100%) by patient characteristics and number of symptoms. On the other hand, the specificity of the new test varied depending on how soon patients were tested after onset of symptoms (100% specificity when swabs were taken on the first day of the symptoms, decreasing to 75% when swabs were taken on or after the third day). The specificity of the new test also increased with increasing body temperature.
CONCLUSION:
The new test seems to provide a cost-effective alternative to real-time RT-PCR for diagnosing H1N1 influenza. However, further testing may be needed to verify the efficacy of the test in different settings and communities.
The laboratory diagnostic tests used for identifying influenza viruses in respiratory specimens differ in their sensitivity and specificity as well as in their commercial availability.1 Additional problems include the amount of time needed to process specimens and the difficulty in distinguishing between different virus types and subtypes (e.g., novel versus seasonal H1N1). Rapid testing, the first step in the influenza A (H1N1) screening process, seems to be of limited value in providing a strain-specific H1N1 diagnosis.2,3 Additionally, a negative rapid test result does not rule out an influenza A (H1N1) virus infection.2,4–8 On April 28, 2009, the World Health Organization (WHO) issued the first revision of the Centers for Disease Control and Prevention protocols for using real-time RT-PCR as a diagnostic test for novel influenza (H1N1). The protocol recommended the use of real-time RT-PCR as a diagnostic test for the pandemic H1N1 influenza virus.9 This test has high sensitivity and specificity, but is expensive and requires technical expertise—drawbacks that may prevent its widespread use in most low- and middle-income countries (LMICs). This highlights the need for an alternative test that can match the sensitivity and specificity of real-time RT-PCR, without most or all of the problems associated with its use. A new kit, Dr. KSU H1N1 RT-PCR kit (hereafter referred to as, simply, the “new test”), has been developed in response to this need. The efficacy of this kit was examined in a pilot study conducted in China (A. Al-Khedhairy, unpub. data). The preliminary results were promising, but a larger, well-designed validation study was still required to make this test medically acceptable for practical use. Hence the current work presented here was designed to assess the diagnostic accuracy of the new test in comparison with the current gold standard, real-time RT-PCR.
PATIENTS AND METHODS
This study included data from patients who attended an H1N1 influenza clinic from 18 to 22 November 2009. This study received the approval of the institutional ethical review board; patient consent was obtained after the study protocol was explained to the patients. They were included in the study only if they had not already received an antiviral treatment. Each patient was given a structured questionnaire to collect the following information: socio-demographic data (age and gender), number of days the patient experienced symptoms before attending the clinic, and symptoms (e.g., fever, rigors, headache, muscle aches, nausea, sore throat, sneezing, cough, and/ or fatigue). Samples from each patient's nasopharynx were collected using nasopharyngeal swabs. All swabs were sent to the molecular biology laboratory at the King Khalid University Hospital for blind testing by both real-time RT-PCR and the new test (described below). Outcomes of each test were entered into Microsoft Access software (Microsoft Corporation, Redmond, WA, USA) and analyzed using MedCalc (Mariakerke, Belgium) and Epi Info software (CDC, Atlanta, GA, USA). The kappa statistic was used to assess the agreement between the results of the two influenza tests (reliability testing). The test characteristics of the new diagnostic kit (e.g., sensitivity, specificity, positive and negative predictive values, and positive and negative likelihood ratios) were also calculated (validity testing), along with corresponding 95% confidence intervals (CIs).
Real-time RT-PCR
High Pure viral nucleic acid kits (Roche Company, Germany) were used to extract nucleic acids. The extraction process is as follows: Viruses, when lysed by detergent and proteinase K, release total viral nucleic acid (NA). Then, in the presence of a chaotropic salt (guanidine HCL), viral NA binds selectively to glass fiber fleece in a special centrifuge tube. The NA remains bound while a series of rapid “wash-and-spin” steps remove contaminating cellular components. Finally, low salt elution removes the NA from the glass fiber fleece. The process does not require NA precipitation, organic solvent extraction, or extensive handling of the NA. Influenza A/H1N1 RNA virus detection was performed using a real-time Ready Influenza A/H1N1 detection kit on a LightCycler 1.2 (Roche) according to the manufacturer's instructions. Nucleic acids were stored at -70°C until tested.2,3 Once genome extraction is complete, this procedure requires approximately 6 hours to amplify 90 extracted samples and produce the final test results.
New test
The new test (Dr. Chip Biotech Company, Taiwan) is designed to simultaneously detect influenza A, influenza B and the novel influenza A virus (H1N1) in a single polymer chip. The kit combines four molecular biological techniques: viral RNA extraction, reverse transcription (RT), polymerase chain reaction (PCR) and Dr. Chip's proprietary nucleic acid hybridization techniques. After the hybridization reactions, the hybridized target DNA is detected by the colorimetric method; the signal is captured and analyzed by Dr. Chip's proprietary imaging device, the Dr. AIM Reader. Once genome extraction is complete, this procedure requires approximately 6 hours to amplify 192 extracted samples and produce the final test results.
RESULTS
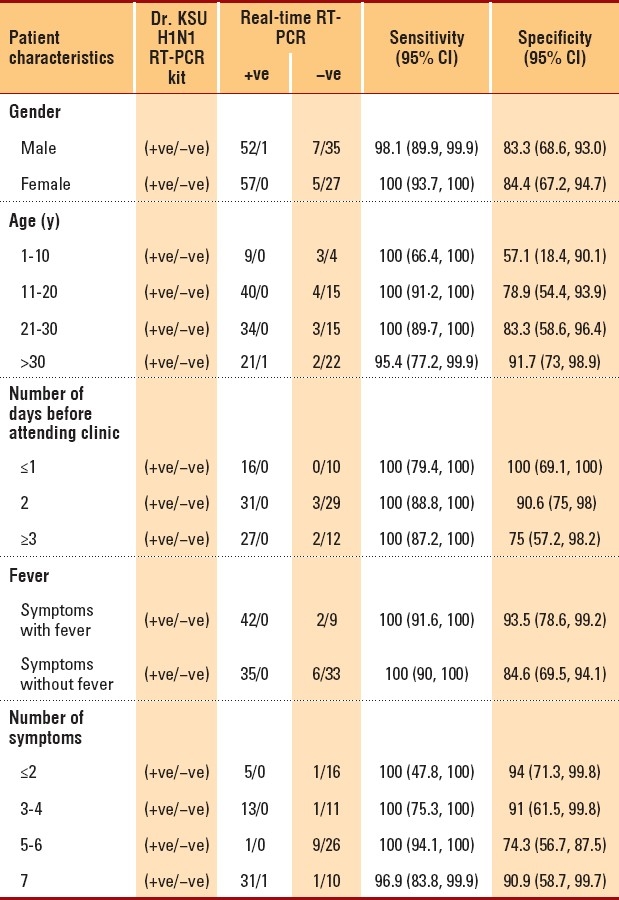
Of the 186 recruited subjects, there were approximately equal numbers of each gender and the mean patient age was 24.9 years (Table 1). Patient body temperatures ranged from 36°C to 40°C. Patients waited for an average of 2.5 days (range, 0-10 days) before attending the influenza clinic. Overall, the prevalence of H1N1 infection in the study population was 60.2% (95% CI, 52.6%-67.3%) (Table 1). A strong agreement was found between the outcomes of the new test and real-time RT-PCR (Table 2) (kappa=0.85; P<.0001). Additionally, the distributions of the two tests were found to be strongly associated (χ2=1376.69; P<.00001), indicating that the new test is highly “reliable.” The characteristics and validity of the new test were reflected by the sensitivity, specificity, and other diagnostic test parameters (Table 3). The sensitivity and specificity of the two tests were further analyzed after separating the data according to patient characteristics and number of symptoms (Table 4). Sensitivity changed only minimally (ranging from 96% to 100%). On the other hand, specificity varied considerably in some categories. For instance, specificity of the new test had an inverse relationship with the number of days that had passed before the patient attended the H1N1 influenza clinic: Among patients who were swabbed on the first day of their symptoms, specificity was 100%; among patients who were swabbed after 3 or more days, specificity decreased to 75%. Additionally, specificity increased with increasing body temperature and age. Among patients who were ≤10 years old, specificity was 57%, but it increased to approximately 94% among patients who were ≥30 years old. Among other variables (e.g., gender), specificity either did not show much variation or was random (as with number of symptoms). Given the time and resources devoted to each test in the molecular laboratory, we calculated that the cost of RT-PCR testing was approximately $25-30 per patient, while the cost of the new test was only $12-14 per patient.
Table 1.
Demography and clinical characteristics.
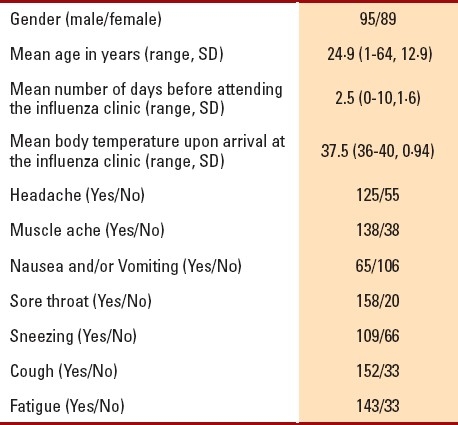
Table 2.
Comparison between Dr. KSU H1N1 RT-PCR kit and realtime RT-PCR.
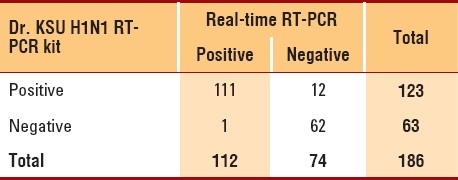
Table 3.
Comparison of diagnostic parameters.
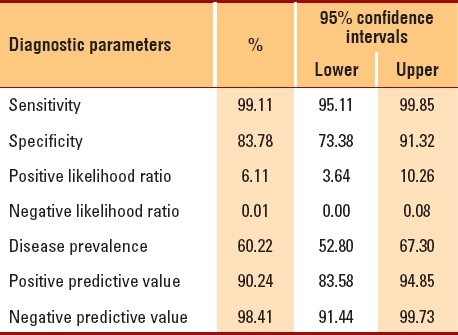
Table 4.
Sensitivity and specificity of the two tests, separated by patient demographics and symptoms.
DISCUSSION
Tracking novel influenza viruses that have the potential to cause pandemics, such as the pandemic (H1N1) 2009 virus, is a public health priority. To achieve this goal, well-validated diagnostic tools that are rapid, sensitive and specific for the detection and tracking of this virus are needed. Real-time RT-PCR has been recommended by both the CDC and WHO.9 This procedure was developed and validated by Pabbaraju et al10 for diagnosis and surveillance of the pandemic (H1N1) 2009 virus. Our new test (Dr. KSU H1N1 RT-PCR kit) appears to be a highly “reliable” alternative to real-time RT-PCR, as demonstrated by both the kappa statistic and the chi-square test. Moreover, with real-time RT-PCR set as the gold standard, the new test showed high “validity,” as reflected by its high sensitivity and specificity. However, while sensitivity changed only minimally in response to infection development and number of symptoms among our patients, specificity changed significantly. Additionally, we found that specificity decreased as the number of days before attending the clinic increased; on the other hand, specificity increased as body temperature increased. Although specificity was shown to vary with age, no definite conclusions could be made, because some categories (e.g., children <10 years) had limited numbers. The variation in specificity that we observed among our patients could be explained by variation in the natural viral load of patients with novel A (H1N1) influenza, an issue that has been debated in recent literature.11,12 To et al11 reported viral loads of pandemic H1N1 virus in respiratory specimens, stool, urine and serum, as determined by quantitative RT-PCR, where respiratory specimens from patients with seasonal influenza were used as historical controls. Among patients with pandemic H1N1 virus infections, peak viral load occurred on the day of symptom onset and declined gradually thereafter. Except in one patient, culturing and RT-PCR detected no virus in respiratory specimens by 5 and 8 days, respectively, after the onset of symptoms. On the other hand, Li et al8 used RT-PCR in a retrospective cohort study involving 145 patients whose specimens tested positive for the matrix and new H1 genes. This study found no correlation between viral load and patient age and number of symptoms. The high sensitivity and specificity of the new test could avoid the false-negative results that have recently been reported for real-time RT-PCR. In fact, Rello et al13 discuss four cases (12.5% of the total sample) where real-time RT-PCR results were negative when patients were admitted to the intensive care unit (ICU), but were positive when the patients were later intubated. On the basis of these findings, the authors conclude that negative results should not exclude influenza A (H1N1) diagnosis. On the other hand, Gimeno and Navarro14 argue that the negative results reported by Rello et al11 cannot be considered true false-negatives, because the samples were not tested in parallel with a different assay, yielding positive results. They add that the optimal sensitivity of RT-PCR and rapid antigen tests is achieved when upper respiratory tract specimens are collected within the first few days after the onset of symptoms, as appeared to be the case for the aforementioned patients. Thus inappropriate sampling, specimen processing and/or suboptimal sensitivity of the PCR assay probably accounted for these negative results. The possibility of false-negative RT-PCR results for influenza A (H1N1) virus in severely ill patients requiring admission to ICUs is a very important issue that must be further investigated. Further, the relatively low specificity expressed when comparing our new test with real-time RT-PCR could, in fact, reflect higher sensitivity of the new test (the reverse of specificity) if the comparison was done in reverse (e.g., with the new test as the gold standard); this would probably avoid most of the false-negative results discussed above. The new test has a sensitivity of 99.1%, a specificity of 83.8%, has the ability to process more samples and is more economical than real-time RT-PCR. Thus the Dr. KSU H1N1 RT-PCR kit appears to be a good alternative for clinical use in detecting pandemic influenza A (H1N1), especially in LMICs, requiring a more cost-effective diagnostic tool. However, further testing may be needed to verify the usefulness of this kit in different settings and communities. During emergency situations, such as the current pandemic situation and other times of mass population demand, the new test is highly capable of providing quick, valid and reliable population-wide results at a very competitive price, with minimum expertise. To further understand the reasons behind the lower specificity of the new test, future research should be designed to investigate the possibility that the false-positive responses seen here are a result of higher sensitivity of the new test in comparison to real-time RT-PCR. Specifically, we recommend that the results of the two tests should be compared for multiple samples collected from the same patients at multiple time points.
Contributors
All the authors contributed to the design of the study and criticized drafts of the report, except for the second author. Since the second author of this manuscript, AAK, was involved in developing the new test kit, his role in the study was limited to performing the new test and in training technicians in the mechanics of testing samples and reading test results and writing the methods section for the new test. AAB and AM were responsible for overall supervision of the study. SS was responsible for data management, statistical analysis and preparation of the tables for the manuscript. AAB, AM, RQ and MO contributed to the interpretation of the results. ASK and HAH were responsible for testing the collected samples in the molecular biology laboratory and writing the real-time RT-PCR section in the methods section. AA, LAA and RQ supervised the data-collection process and monitored collection and handling of the nasopharyngeal swabs. AAB, AM, SS and RQ wrote the first draft of the report. AAB was responsible for collecting the inputs of all the co-authors and redrafting the manuscript accordingly.
Acknowledgments
This project was funded by the Standing Committee of Epidemic Control, College of Medicine and King Khalid University Hospital, King Saud University, Riyadh, Saudi Arabia.
REFERENCES
- 1.Uyeki TM. Influenza diagnosis and treatment in children: A review of studies on clinically useful tests and antiviral treatment for influenza. Pediatr Infect Dis J. 2003;22:164–77. doi: 10.1097/01.inf.0000050458.35010.b6. [DOI] [PubMed] [Google Scholar]
- 2.Atlanta, GA: US Department of Health and Human Services, CDC; 2009. [Last accessed on 2009 Dec 2]. CDC. Interim guidance for the detection of novel influenza a virus using rapid influenza diagnostic tests. Available from: http://www.cdc.gov/h1n1flu/guidance/rapid_testing.htm . [Google Scholar]
- 3.CDC. Evaluation of rapid influenza diagnostic tests for detection of novel influenza A (H1N1) virus - United States. Morb Mortal Wkly Rep. 2009;58:826–9. [PubMed] [Google Scholar]
- 4.Ginocchio CC, Zhang F, Manji R, Arora S, Bornfreund M, Falk L, et al. Evaluation of multiple test methods for the detection of the novel 2009 influenza A (H1N1) during the New York City outbreak. J Clin Virol. 2009;45:191–5. doi: 10.1016/j.jcv.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.CDC. Performance of Rapid Influenza Diagnostic Tests During Two School Outbreaks of 2009 Pandemic Influenza A (H1N1) Virus Infection-Connecticut, 2009. Morb Mortal Wkly Rep. 2009;58:1029–32. [PubMed] [Google Scholar]
- 6.Uyeki TM, Presad R, Vutotich C. Low sensitivity of rapid diagnostic tests for influenza. Clin Infect Dis. 2009;48:e89–92. doi: 10.1086/597828. [DOI] [PubMed] [Google Scholar]
- 7.CDC. Hospitalized patients with novel influenza A (H1N1) virus infection---California, April--May, 2009. Morb Mortal Wkly Rep. 2009;58:536–41. [PubMed] [Google Scholar]
- 8.Kok J, Blyth CC, Foo H, Patterson J, Taylor J, McPhie K. Comparison of a Rapid Antigen Test with Nucleic Acid Testing During Co-circulation of Pandemic Influenza A/H1N1 2009 and Seasonal Influenza A/H3N2. J Clin Microbiol. 2010;48:290–1. doi: 10.1128/JCM.01465-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.WHO. CDC protocol of real time RTPCR for swine influenza A(H1N1) 2009. Apr 28, [Last accessed on 2009 Dec 4]. Revision 1(30 April 2009). Available from: www.who.int/csr/resources/publications/swineflu/CDCrealtimeRTPCRprotocol_20090428.pdf .
- 10.Pabbaraju K, Wong S, Wong AA. Design and validation of real-time reverse transcription-PCR assays for detection of pandemic (H1N1) 2009 virus. J Clin Micro. 2009;47:3454–60. doi: 10.1128/JCM.01103-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.To KW, Chan KH, Li IWS. Viral load in patients infected with pandemic H1N1 2009 influenza A virus. J Med Virol. 2010;82:1–7. doi: 10.1002/jmv.21664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li IW, Hung IF, To KK, Chan KH, Wong SSY, Chan JF, et al. The natural viral load profile of patients with pandemic swine-origin influenza A H1N1 2009 (pH1N1) and the effect of oseltamivir treatment. Chest. 2010 doi: 10.1378/chest.09-3072. [In Press] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rello J, Rodríguez A, Ibañez P, Socias L, Cebrian J, Marques A, et al. The H1N1 SEMICYUC Working Group: Intensive care adult patients with severe respiratory failure caused by influenza A (H1N1)v in Spain. Crit Care. 2009;13:R148. doi: 10.1186/cc8044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gimeno C, Navarro D. Real-time reverse-transcription PCR in the diagnosis of influenza: A (H1N1)v in intensive care unit adult patients. Crit Care. 2009;13:428. doi: 10.1186/cc8164. [DOI] [PMC free article] [PubMed] [Google Scholar]


