Abstract
Understanding how genes and the environment interact to shape phenotypes is of fundamental importance for resolving important issues in adaptive evolution. Yet, for most model species with mature genetics and accessible genomic resources, we know little about the natural environmental factors that shape their evolution. By contrast, animal species with deeply understood ecologies and well characterized responses to environmental cues are rarely subjects of genomic investigations. Here, we preview advances in genomics in aphids and waterfleas that may help transform research on the regulatory mechanisms of phenotypic plasticity. This insect and crustacean duo has the capacity to produce extremely divergent phenotypes in response to environmental stimuli. Sexual fate and reproductive mode are condition-dependent in both groups, which are also capable of altering morphology, physiology and behavior in response to biotic and abiotic cues. Recently, the genome sequences for the pea aphid Acyrthosiphon pisum and the waterflea Daphnia pulex were described by their respective research communities. We propose that an integrative study of genome biology focused on the condition-dependent transcriptional basis of their shared plastic traits and specialized mode of reproduction will provide broad insight into adaptive plasticity and genome by environment interactions. We highlight recent advances in understanding the genome regulation of alternative phenotypes and environmental cue processing, and we propose future research avenues to discover gene networks and epigenetic mechanisms underlying phenotypic plasticity.
Keywords: adaptive plasticity, alternative phenotypes, condition-dependent transcription, epigenetics, gene regulation, genome-by-environment interaction, polyphenism
Over the last decade, there has been an increasing awareness of the importance of phenotypic plasticity in structuring ecological interactions (Agrawal et al. 1999; Pohnert et al. 2007), in tolerating environmental fluctuations (Tollrian and Harvell 1999), in adaptive evolution, ecological speciation (Miner et al. 2005; Pigliucci 2005; West-Eberhard 2003), and populations persistence and extinction in changing environments (Chevin et al. 2010). Yet, our understanding of the molecular and cellular process by which a plastic response is triggered by environmental cues is limited (Aubin-Horth and Renn 2009). The concept of phenotypic plasticity predates the modern evolutionary synthesis. Importantly, early researchers discovered that changes in the phenotype due to environmental effects result in context dependent changes in fitness (e.g., Baldwin 1896; Woltereck 1909). However, the role of plasticity in adaptive evolution remained somewhat controversial for much of the last century (Sarkar 2004). Beginning in the 1980’s, renewed interest in plasticity motivated empirical studies, particularly in plants, and the development of theoretical models describing the evolutionary dynamics of phenotypic plasticity (Via et al. 1995; Berrigan and Scheiner 2004). These models approached adaptive plasticity from a number of different perspectives including quantitative genetics and optimization models. Three important questions emerged: 1) Does substantial genetic variation for plasticity segregate in natural populations?, 2) Is phenotypic plasticity an adaptation to divergent selective regimes and 3) Do “plasticity genes” regulate the environmentally dependent expression of plastic traits and at what cost. The first two issues have largely been resolved by the numerous studies demonstrating heritable variation and adaptive value for plasticity (Scheiner 1993; Via et al. 1995; Charmantier et al. 2008). However, the existence of plasticity genes is less well demonstrated. Genome-wide association studies for detecting genes linked to phenotypes strongly suggest that complex traits are governed by many loci (Flint and Mackay 2009); therefore, plasticity genes are most likely to be global regulators of gene products. A general understanding of developmental genetics suggests that a broad array of regulatory mechanisms involving epistatic interactions among genes (Carlborg and Haley 2004), small RNA molecules (Behura 2007), transcriptional factors or epigenetic processes, may potentially be co-opted to mediate phenotypic plasticity.
A significant outstanding issue is the relationship between genome structure and adaptation. There is some evidence that co-expressed genes tend to show a non-random pattern of co–localization within eukaryotic genomes (Hurst et al. 2002; Williams and Bowles 2004). Yet, how selection and environmental context influence the physical arrangement of the multiple genes and regulatory elements that underlie polygenic adaptive traits remains a poorly explored question (but see Gutteling et al. 2007). In the context of phenotypic plasticity, the relationship between environment and genome structure may be a particularly relevant issue, since context dependent fitness may be the direct result of the interaction between environmental effects and a suite of multiple genetic factors. An understanding of the genetic and developmental basis of plasticity is also critical for the application of recent theoretical models that allow for context-dependent evolution (e.g., Rice 2002, 2004; Wolf et al. 2004). The continued development of ecological genomic model species such as Arabidopsis and arthropods, including aphids and Daphnia, with advanced genomic tools and well understood ecologies is facilitating rapid progress in these areas. For example, recombinant inbred lines have been developed in Arabidopsis to examine the costs of plasticity (Callahan et al. 2005), genome-wide patterns of gene expression have been analyzed (Schmid et al. 2005), and a growing number of Quantitative Trait Loci (QTL) studies in explicit environmental contexts are being been conducted (Weinig and Schmitt 2004). The emerging picture from studies in Arabidopsis is that many QTL have environment specific effects (reviewed in Mitchell-Olds and Schmitt 2006). QTL-environment interactions appear to be common and highly variable (Ungerer et al. 2003; Juenger et al. 2005) and epigenetic variation can have a dramatic impact on the plasticity of ecologically relevant traits (Johannes et al. 2009).
The traditional model organisms (i.e., Escherichia, Saccharomyces, Arabidopsis, Caenorhabditis, Drosophila, Danio, and Mus) were selected for their utility in developmental and cellular biology, and genetics. Unfortunately, these systems often lack significant biological context outside of the laboratory. Yet, one of the ultimate goals of biology is to understand how genome structure and function evolve in response to environmental change and how genetic constraints limit the natural distribution and abundance of species and populations. In this paper, we review recent research on the genomics and genetics of phenotypic plasticity in two groups of invertebrates, aphids and Daphnia. These organisms are model systems for integrative studies on phenotypic plasticity spanning levels of organization from gene to phenotype to ecological context.
Aphids and Daphnia as Model Systems for Studies on Phenotypic Plasticity
Aphids and Daphnia show several discrete alternative phenotypes produced in response to environmental changes. This phenomenon termed polyphenism is a special case of phenotypic plasticity defined as “the ability of organisms with the same genotype to develop two or more distinctly different alternative phenotypes without intermediates” (Nijhout 1999). As developed in the next section, aphids and Daphnia share the ability to generate, from the same genetic background, alternative forms differing in morphology, physiology, ecology and behavior. Also, their shared cyclically parthenogenetic life cycle (i.e., regular alternation of clonal and sexual generations) allows propagation by cloning, selfing, or outcrossing, therefore providing excellent opportunities for genetic analysis. Clonal lineages allow the side-by-side measurement of phenotypes produced by the same genome (in a fixed genetic background) and partitioning of the genetic and environmental components of phenotypic plasticity. Their laboratory attributes are as compelling as those of other model systems; they have short generation times when reproducing asexually (∼10 days in most aphid and Daphnia species at 20 °C), high fecundity (up to 100 daughters from a single aphid and 10-100 per clutch for a single Daphnia), they produce diapausing propagules for long-term storage, and they are easy to maintain. In addition, all life stages can be observed and studied in nature, and populations are easily defined and readily sampled in large numbers. Aphids and Daphnia have been recent targets for genomic studies. In each group, a complete draft genome sequence is now available for a representative species (the pea aphid Acyrthosiphon pisum and the waterflea Daphnia pulex). Our scientific communities have produced genomic resources for high-throughput molecular biology, for functional genomic studies and databases that initiate important research programs to decipher the genetic, molecular, cellular, physiological basis of several plastic traits.
The microcrustacean Daphnia has been subject to over a century of study in the areas of limnology, life history, physiology, nutrition, predation, parasitology, toxicology, and behavior. There is a well-resolved phylogenetic framework for the genus (Adamowicz et al. 2009; Colbourne and Hebert 1996) allowing multiple levels of comparative genomic investigation. Daphnia exist in diverse environments (e.g., permanent lakes and temporary ponds) spanning a wide range of ecological conditions, including heavily damaged ecosystems resulting from human activities. As a result, Daphnia are notable for rapid adaptive evolution (Cousyn et al. 2001; Fisk et al. 2007) in response to changing environmental conditions. This ecological diversity provides a unique opportunity to ask if Daphnia evolve to meet environmental challenges the same way in independent lineages (Colbourne et al. 1997; Pfrender et al. 2000; Scoville and Pfrender 2010).
Aphids, which comprise ∼4,400 known species worldwide, have been studied intensively mainly because of their economical impacts on agriculture, reducing considerably crop yields and transmitting a plethora of plant viruses. Many areas of research have been explored in this insect group from fundamental works in evolutionary biology, ecology, symbiosis, developmental and reproductive biology to more applied studies on pest control via the use of chemicals, resistant plants and natural enemies (Dixon 1998). As for Daphnia, a solid phylogenetic framework exists in aphids and advanced genomic resources are being accumulated in multiple aphid lineages for use in comparative and evolutionary genomics (Ollivier et al. 2010; Sabater-Munoz et al. 2006). Aphids also inhabit a wide range of ecosystems from polar to tropical, interact with a huge diversity of biotic (plants, micro-organisms, natural enemies, human activities) and abiotic factors (climate, chemicals, pollutants) and evolve rapid adaptive responses (e.g. insecticide resistance, breakdown of plant resistance), allowing studies on the evolution of biological adaptations, including adaptive phenotypic plasticity (Le Trionnaire et al. 2008; Tagu et al. 2008; Brisson 2010).
Alternative Phenotypes in Aphids and Daphnia—A Case for Comparative Genomic Research
Aphids and Daphnia share a rare suite of multiple alternative phenotypes allowing them to cope with environmental changes and to anticipate deteriorating habitats and resources. A successful approach to questions on the origin and maintenance of adaptive traits is to study independently derived features through comparative analysis (Brooks and McLennan 1991). The spectacularly plastic phenotypes in Daphnia and aphids are excellent examples of independent convergence and provide an opportunity to examine the commonalities of affected developmental pathways in depth. The range of phenotypes and ecological context within an aphid or daphniid species often rivals the variation found between distantly related species (Petrusek et al. 2009), forming ideal empirical systems for a comparative functional genomic approach to plasticity.
Alternative Phenotypes Involved in Reproduction
The dominant reproductive mode in animals is sexual reproduction. Alternative reproductive modes include obligate asexuals, which are fairly rare, and cyclical parthenogens. Organisms with cyclic parthenogenic life histories, including aphids and waterfleas, (De Meester et al. 2004), capitalize on the advantages of the two reproductive modes by alternating between sexual and asexual reproduction in response to environmental conditions. The shift between asexual and sexual development in aphids is triggered by photoperiodic changes: clonal aphids perceive the increase in night lengths via photoperiodic photoreceptors at the anterior dorsal region of the protocerebrum and respond over a transgenerational process by producing sexual forms (Tagu et al. 2005). In addition, clonality is associated with viviparity while sexuality is linked with oviparity (Le Trionnaire et al. 2008). Viviparous forms are frost-susceptible while oviparous ones lay cold-resistant and diapausing eggs that hatch the following spring. Similarly, during the growing season, Daphnia produce sizable broods of genetically identical daughters that directly develop through juvenile instars into adults (Berg 1934). In Daphnia, as in aphids, environmental factors that include photoperiod (Banta and Brown 1939; Stross and Hill 1965), crowding (Carvalho and Hughes 1983), starvation and temperature (Alekseev and Lampert 2001) stimulate the switch from producing ameiotic diploid eggs to a pair of meiotic haploid eggs requiring fertilization for development. Here, sexuality is also linked with oviparity. In a state of diapause, the embryos are encapsulated within a protective structure called an ephippium, which is resistant to freezing and desiccation, until environmental cues trigger development (Davison 1969; Pfrender and Deng 1998). Importantly, while the switch in reproductive modes and the termination of diapause in Daphnia is largely precipitated by environmental cues, there is substantial genetic variation within and among populations and species for these plastic traits (De Meester and Dejager 1993; Deng 1996; Pfrender and Deng 1998; Yampolsky 1992). The same applies to aphids which show, within a single species, co-existing lineages with varying levels of investment into the sexual phase i.e., from complete loss of sex to regular alternation of clonal and sexual phase with all intermediates in between (Simon et al. 1999, 2002).
Arguably, a more remarkable phenotypic transformation related to reproduction is the switch from producing genetically identical daughters to genetically identical sons. Males differ from females in morphology, behavior, responses to stimuli and life history. Moreover, gene expression studies of insects suggest that 12-32% of the genes in Drosophila transcriptomes are sex biased (Zhang et al. 2007) and that gamete formation accounts for approximately 30% of the transcriptional differences between the sexes (Parisi et al. 2004; Ellegren and Parsch 2007). However, unlike Drosophila, sex determination in Daphnia is triggered by environmental cues in a fashion similar to the switch in reproductive tactics. Although the same environmental factors may specify deteriorating conditions for the start of sexual reproduction (Banta and Brown 1939; Yampolsky 1992), the signaling pathways are seemingly independent. Daphnia populations exhibit substantial variation in the onset of male production and the onset of meiotic oogenesis, including isolates that either fail to reproduce sexually or are unable to produce male offspring (Innes and Dunbrack 1993; Innes and Singleton 2000). Recent studies implicate endocrine cascades at the head of the Daphnia sex-determination pathway, by the action of the juvenile hormone methyl farnesoate in signaling male production (Olmstead and LeBlanc 2003). Interestingly, the same pathway also signals hemoglobin production when D. pulex is starved of oxygen – another significant plastic trait. Non-male producing clonal isolates have varying defects along the signaling pathways, based on observations that not all non-male producers fail to also produce hemoglobin (Rider et al. 2005). Obviously, genetic mapping studies using multiple isolates that differ in their combined responses to hypoxia and environmental stress signals will be necessary to disentangle the genetic interactions for these traits.
In aphids, there also exists a difference in the cues inducing males and those inducing meiotic oogenesis. Male production requires specific photoperiodic and/or temperature regimes leading to the loss of one of the two sexual chromosomes (in aphids, sex determination is XX:XO female:male). Sex determination seems to depend on the concentration of juvenile hormone (JH) with high titers leading to females and lower ones to males (Hales and Mittler 1987). As for Daphnia, aphids encompass genotypes differing in their capacity to produce males, this polymorphism being controlled by a limited number of genes (Simon et al. 1999). A comparative approach of gene regulation associated with male production within the same genotype or between genotypes with varying investment in the male function will certainly help to decipher the cascade of molecular and cellular changes from perception of the environmental signal by the neural system to its transduction via the endocrine system to the reproductive track.
Alternative Phenotypes Involved in Defense or Dispersal
Aphids and Daphnia also show spectacular morphological changes, such as individuals with or without wings in aphids (Figure 1), and forms with or without defense structures against predators in both aphids and Daphnia (Figure 2). These plastic morphologies, forming complex reaction norms, are adaptive responses in anticipation of changing or deteriorating environmental conditions. Most aphid individuals are wingless, feeding on plants and allocating massively in reproduction by viviparous parthenogenesis. However, aphids are also found as winged phenotypes able to dispersal and occupy free habitats. Sexual forms, or their precursors, produced in the autumn are usually winged, allowing migration to mating sites (e.g., a specific host-plant where sexual reproduction takes place) and preventing inbreeding costs. In some species, males can be winged or wingless depending on the genotype. This variation is controlled by a biallelic locus (Braendle et al. 2006; Brisson 2010). Winged forms are also produced during the parthenogenetic phase of the aphid life-cycle. In this case, wings are induced when colony density is high and resources limiting, or when plant quality is deteriorating. Interestingly, variation in wing production in asexual females is linked to that of wing male phenotypes, suggesting that the same set of genes could be involved in both phenotypic plasticity and polymorphism (Braendle et al. 2005; Brisson 2010). Since the proximal cue for wing plasticity is the perception of individual movements within the colony through antennal contacts (Kunert et al. 2005), winged aphids are also produced in greater numbers when colonies are attacked by predators (e.g., ladybird beetles) or parasites (e.g., parasitoid wasps). Also, like other social insects such as bees, ants and termites, a few aphid species show caste determination with soldiers and normal forms. Soldier morphs possessing enlarged prehensile forelimbs and frontal horns, occur in less than 1% of aphid species and in only two families, the Hormaphididae and Pemphigidae (Stern and Foster 1996). Phylogenetic analyses indicate that soldiers have independently evolved a minimum of 6-9 times. Soldier morphs are sterile and show defense and attack behaviors to protect the colony against predators. In the Asian aphid Tuberaphis styraci, the colony develops within a gall on the tree Styrax obassia and soldier morphs differentiate when colony reaches high density. These soldiers are aggressive against enemies, by inserting their mouthpart inside the intruder body. Attacks are often lethal for the predator, even if its body size exceeds 1,000 times that of the aphid.
Figure 1.
Inducible wing polyphenism in the vetch aphid Megoura viciae (A) wingless form produced in favourable conditions (e.g. low density). (B) winged form produced in deteriorating conditions (e.g. crowding). Photos by Bernard Chaubet. (This figure appears in color in the online version of Journal of Heredity.)
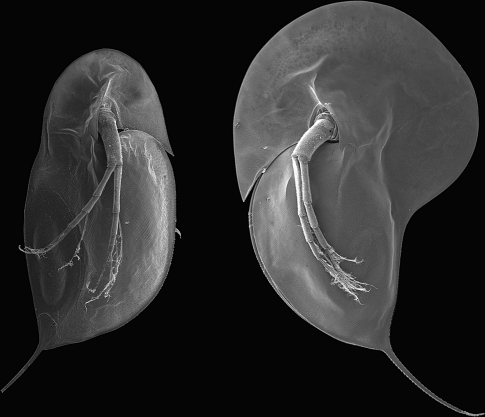
Figure 2.
Inducible defense structures in Daphnia longicephala. Identical genotypes showing the effect of waterborne chemical cues from predators (kairomones). Non-induced morphology on the left and induced morphology on the right. Image by Dr. Christian Laforsch.
The developmental program underlying the determination of alternative phenotypes in aphids is far from well understood, but this biological system attracts increasing attention from evo-devo researchers. Comparative analysis of sexual and asexual development in the pea aphid has been conducted using anatomical, morphological and histological approaches (Braendle et al. 2003; Miura et al. 2003). This research indicates that sexual and parthenogenetic embryogenesis progress through two strongly contrasted pathways (Le Trionnaire et al. 2008), from oocyte determination (meiotic oogenesis in sexual females vs. mitotic division in asexual ones) to body, reproductive, sensorial and neural development. The developmental pathways of winged or wingless phenotypes under the right conditions (crowded vs. uncrowded, respectively) have been less well studied. A recent anatomical analysis clearly indicated that embryos and larvae, in both winged and wingless individuals, develop in their early stages wing primordia responsible for wing formation in insects (Ishikawa and Miura 2007; Ishikawa et al. 2008). This work showed that the developmental cascades that produce wings and flight muscles are switched on during the first stages of future winged form, while the degeneration process is launched in the early development of future wingless form. The default genetic program in aphids is thus wing development during embryogenesis (as in other insects). This observation also suggests differences in gene expression patterns in the wing primordial in early stages and responsible for the wing plastic response in aphids. Interestingly, winged and wingless phenotypes not only differ in morphology but also in other traits, such as reproduction, behavior, and sensory system (Dixon 1998).
Daphnia are famous for their phenotypic plasticity in defensive traits. These small organisms exhibit an astonishing repertoire of inducible defenses by modifying morphological, physiological or behavioral traits (Tollrian and Dodson 1999). Morphological changes in most cases lead to enlargements of different body parts, including a marked elongation of the tail spine or the head and the development of spiky or curved helmets or dorsal crests, or the size of the compound eye. These changes often interfere with prey capture or consumption and have a demonstrable fitness benefit (Tollrian 1995; Petrusek et al. 2009).
Physiological traits lead to changes in life-history parameters, including diapausing egg induction that allows the prey to temporally avoid predators, or shifts in resource allocation between somatic growth and reproduction (Stibor and Luning 1994; Latta et al. 2007). These defenses usually interfere with prey encounter or recognition if the predators use visual cues to target larger Daphnia (e.g., fish) or with prey handling if the predators use tactile cues and select smaller individuals (e.g., invertebrates). Behavioral defenses such as elevated alertness, modified swimming behaviors, the formation of swarms or diel vertical migration may reduce encounters or allow escape.
In nature, most prey species are threatened simultaneously by a variety of predators with different prey preferences and different hunting strategies. Hence, many prey species have evolved several inducible traits which allow a flexible response to a changing predator regime. Among Daphnia species, protective mechanisms differ even against the same predator. For example, D. pulex generates neckteeth primarily in the second instar in the presence of Chaoborus larvae (the phantom midge, a voracious predator of freshwater zooplankton), whereas D. cucullata shows helmet formation during all life stages when faced with this predator. This varied response is presumably due to differing life-stage specific susceptibility to Chaoborus predation. The smaller of the two species, D. cucullata, is within the prey-size range of Chaoborus in all instars, while the larger species, D. pulex, is vulnerable only during juvenile instars. Furthermore the helmets in the small D. cucullata form a “multitool” against different predators (Laforsch and Tollrian 2004), while neckteeth are specific defenses against a single predator species. The two morphological defenses, exaggerated helmets and dorsal neckteeth, are repeatedly found in independent lineages across the genus (Colbourne et al. 1997).
The fitness benefit of inducible defenses depends on the lag phase between the onset of the ecological pressure promoting the utility of the induced phenotype and its formation. This is true for wing induction in aphids as well as for helmet and neckteeth formation in Daphnia. Mechanisms which shorten lag phases have been reported in the form of maternal induction of defenses, where neonates already hatch with preformed defenses or enhanced response to environmental cues, and finally reach stronger defense expressions (Agrawal et al. 1999). Transgenerational inductions are beneficial adaptations in situations where the maternal environment is a good predictor of offspring environment. This is the case for wing induction in aphids, where mother's environmental experience influences the phenotype of her offspring, as she contains already developing oocytes and embryos of the two next generations (i.e., telescoping of generations). Different inducible traits vary in their effectiveness, associated costs, and also in the response time between induction and formation. The induction of behavioral traits might generally be faster than the growth of morphological traits, and shifts in life-history traits are likely to be slowest. Thus, a combination of different inducible trait systems could help to reduce the initial lag phases of each until effective defenses are formed. While transgenerationally induced defenses are adaptations to long-term fluctuations in the environment, reversibility of induced traits is beneficial if environmental changes are likely to occur within the life time of an organism (Gabriel et al. 2005). Most behavioral and morphological traits are reversible, while the slower forming life-history changes often are not.
Despite the abundance of research on morphological plasticity, the underlying genetic and cellular processes are not well understood (Tollrian and Leese 2010). In Daphnia, embryonic and juvenile induction occurs, corresponding to the developmental stage where the defence is needed. While it is documented that chemical cues (kairomones) released by the predators are relevant for the induction, neither signal perception nor transduction have been analysed. Formation of helmets in Daphnia can also be induced by sublethal concentrations of some pesticides, including carbaryl and endosulfan (Hanazato 1991; Hanazato 1992) that affect the transmission of nerve impulses in vertebrates and invertebrates. These chemicals may act as acetylcholine esterase inhibitors, resulting in a permanent activation of synaptic activities that stimulate the neurosecretory release of hormones involved in the formation of morphological changes. The involvement of the nervous system in the regulation of phenotypic plasticity has been established for morphological changes in Daphnia (Barry 2002). Stimulation of the cholinergic neurotransmitter system with acetylcholine agonist leads to an increased formation of neckteeth. The morphologically plastic tissues in Daphnia are characterized by endopolyploid cells which are supposed to act as developmental control centers governing the shape of the defensive traits (Beaton and Hebert 1997). Thus, a working hypothesis is that a concentration gradient of hormones in the hemolymph of the animal may be responsible for enhanced cell division rates in the target tissue when the titer exceeds a threshold value. Observations favouring this hypothesis include the number and distribution of polyploid cells in Daphnia, which are strongly correlated to the size and the form of the plastic feature across several species. Additionally, mitotic activity in the cephalic epidermis of a defensive trait has been found to be elevated in regions surrounding these polyploid cells, suggesting the release of a mitogen which triggers the development of the modified trait.
The examples above show that aphids and Daphnia combine extensive phenotypic plasticity with high evolutionary flexibility in terms of genetic responses to changes in ecological conditions. Such a pattern probably arises from the combined effects of long-lived clones with short individual generation times. These high levels of phenotypic plasticity could be explained by the fact that individual genotypes (genetic clones) often live for a long time (for one to many years). As a result, the environment becomes fine-grained at the level of the genotype, resulting in selection for phenotypic plasticity tracking seasonal changes in environmental conditions (De Meester et al. 2004).
Genomic Content and Structure—Early Clues on the Regulatory Mechanisms of Phenotypic Plasticity
Phenotypic plasticity is a consequence of environmental cues that are perceived during ontogeny and the capacity of the individual to integrate cues that influence development. This process can be divided into three main phases: i) the perception of an environmental signal (input), ii) signal transmission and transduction towards the targeted organs, tissues or cells, and iii) differentiation of the induced phenotype (output) by modifying genetic developmental programs. In animals, these steps are clearly dependent on neuro-endocrine regulations (detection, transduction) and other gene expression regulators (output). Discovering these genes and pathways and elucidating their network relationships is a formidable challenge.
Efforts that began in years 2002–2003 by two international research consortia have produced important genomics resources that facilitate molecular, genetic, evolutionary and ecological studies in aphids (www.aphidbase.com) and Daphnia (Colbourne et al. 2005; Legeai et al. 2010a). The first descriptions of the genome structures and gene contents from draft genome sequence assemblies are being published (The International Aphid Genomics Consortium 2010; Colbourne et al. 2011). The 530 Mb genome of A. pisum is assembled into 22000 scaffolds (contiguous DNA fragment reads) covering 87% of the total genome, whilst the modest 200 Mb genome of D. pulex is assembled into 5,000 scaffolds representing 80% of the total nuclear content. The process of finishing these genome projects, by filling gaps and reducing the scaffold numbers to match 4 haploid chromosomes for the pea aphid and 12 chromosomes for Daphnia is underway.
A most fascinating shared feature of these two divergent genomes is their large gene counts. More than 30000 genes are predicted in both species by automated gene calls and by the empirical annotation of genes via their detected transcripts in large scale cDNA sequencing projects and/or whole-genome tiling path microarray studies. These tallies are double the number of genes identified in the current set of sequenced arthropods, such as Drosophila (fruitflies), Anopheles (mosquitoes), Apis (honey bees), Tribolium (flour beetles), Bombyx (silkmoths) or Nasonia (minute parasitic wasps). A closer look at the gene sets indicate a history of gene duplication that accounts for 50% of their gene inventories, which is twice the fraction of duplicate genes among Diptera (Gilbert 2008). Moreover, empirical annotation of the Daphnia genome suggests that more than 8,000 protein coding genes remain undescribed, likely because of their lack of homology to proteomes of other species and their condition specific expression, adding to the already large fraction of lineage specific genes in the two genomes. It is still too early for explanations as to why these two cyclical parthenogens — which are also highly plastic in their responses to environmental changes – should have an independent history of elevated gene amplification. Is this commonality simply a consequence of their shared unique life-cycles that have periodic effects on the effective population sizes of aphid and Daphnia populations? Cyclical parthenogenesis exposes the genome to a more dynamic interplay between selection and drift, which may facilitate the fixation of duplicates (The International Aphid Genomics Consortium 2010). Alternatively, does the genome structure and content reflect an expansion of gene functions and regulations for developmental and behavioral plasticity? As described above, one critical feature of an adaptively plastic phenotype is a condition-dependent specificity. In Daphnia, gene duplicates often have divergent and condition-dependent regulation provocatively suggesting that there may be a direct connection between the maintenance of a large number of duplicate genes and plasticity. For certain, the process of acquiring new genes by duplication is ongoing and has persisted for long evolutionary time in both lineages. Most duplicates in the Daphnia genome are locally arrayed in tandem and therefore are not a consequence of punctuated whole genome duplication events (Colbourne et al. 2011). The nucleotide sequence divergence among duplicate genes suggests that their birth and death rates are more-or-less constant and three times greater than the fastest evolving insect genome (Drosophila; Colbourne et al. 2011). At present, the scattering of aphid genomic fragments into more than 22000 pieces impairs similar conclusions on genome history. However, for both species, additional genome sequence data from phylogenetically related taxa will help determine when runaway gene duplication events began. If lineage specific gene family expansions began at the root of the aphid and waterflea (Cladocera) species radiations, then a stronger case will be made for the link between their genome structures and the shared biology of these two organisms. Nevertheless, a first-pass look at shared gene family expansions and loss produces valuable hypotheses for detailed investigations.
From a total of 27,500 identifiable clusters of homologous genes among 10 insects and a Chelicerate, Daphnia and aphid share 5,436 clusters (Gilbert 2008); 119 gene clusters are uniquely found in our two focal species. These exclusively homologous gene clusters represent obvious candidates for investigations into their roles in the biology of both Daphnia and aphids. Interestingly, a number of homologous gene clusters that are shared with well characterized model systems, like Drosophila, are often found amplified in our insect-crustacean duo compared to those of other arthropods. For example, 78 clusters containing genes that function in chromatin assembly, microtubule formation and mitosis, mitosis kinases, RNA polymerase and binding, plus transcription initiation and regulation, all have elevated gene counts in aphids and Daphnia (on the order of 2-4 times the average number in insects. Clearly, DNA and RNA management genes have jointly diversified, which may be an important clue for understanding mechanisms for altering phenotypes.
The production of active proteins relies on several steps that can be all subjected to regulation: chromatin accessibility (epigenetic marks), RNA transcription, RNA stability and degradation, protein translation, protein stability and degradation and protein modifications. Among the several types of small non-coding RNAs, microRNAs are small double-stranded RNAs encoded as precursors by the genome of eukaryotes. They join a nucleo-proteic complex and bind to targeted mRNAs by base complementarities. The formation of mRNA/microRNA duplex leads to either degradation or inhibition of translation of the targeted mRNAs (Carthew and Sontheimer 2009). Using available genomes, we annotated the gene precursors of microRNAs in A. pisum (Legeai et al. 2010b) and in D. pulex (Colbourne et al. 2011). We found 161 mature aphid microRNAs by combining homology-based, high throughput sequencing and genome scanning methods. This number is greater than counts in other sequenced insects, save Drosophila (Gerlach et al. 2009). By contrast, only 50 mature Daphnia microRNAs are identified, which is the smallest recorded number among the sequenced arthropods. All microRNA loci found thus far in D. pulex have identifiable insect orthologs. By conducting microarray experiments on the pea aphid, we compared microRNA expression profiles among different ecological and developmental conditions. Interestingly, five microRNAs were differentially expressed when comparing parthenogenetic and sexual females produced by the same aphid clone but under different photoperiodic regimes (Legeai et al. 2010b). Three of these microRNAs have unknown functions, whereas mir34 is known to be regulated in Drosophila by ecdysone and juvenile hormones (Sempere et al. 2002). This result suggests that the transduction of the photoperiod signal might involve microRNAs in the pea aphid.
Epigenetic modifications — that modulate chromatin structure and therefore controlling access of the transcriptional machinery to genes — may be an important determinant of phenotypes in the pea aphid and Daphnia. Both species have all three Dnmt gene subfamilies for DNA methylation, and are therefore likely to share the same epigenetic control system preserved in other eukaryotes (Werren et al. 2009). Dnmt1 is the main enzyme for the maintenance of cytosine methylation. Whereby Daphnia has a single copy of this gene, the pea aphid has two copies. Two copies of Dnmt1 are also found in the genome of the honey bee, which has an impressive epigenetic repertoire (Weinstock et al. 2006; Kutsukake et al. 2008). Both the pea aphid and Daphnia have single copies of Dnmt2 and Dnmt3, which typically methylate when no 5mC are found on either DNA stands. Recently, Dnmt2 methyltransferase was shown to have a higher level of expression in aphids that experienced crowding, the main cue for wing morph production (Walsh et al. 2010). The methylation status for a gene encoding a JH binding protein is different between winged and unwinged morphs, suggesting that epigenetic modifications of DNA might regulate hormone availability and dispersal polyphenism.
As the two research communities pursue functional genomic experiments to identify shared genetic pathways between aphids and Daphnia responding to environmental cues, the search for phenotypic plasticity genes will narrow towards candidate loci for genetic confirmation of their effects. Complicating this effort is the observation that a large fraction of the total gene set in these two species is composed of orphans; 36% of Daphnia genes and approx 40% of aphid loci have no known sequence homology to proteins from other species. Experiments using whole-genome tiling-path microarrays and EST sequencing of RNA produced under defined environmental conditions suggest that these unknown genes are often condition specific in their expression and may be expressed only under ecological situations that are unique to the organism (Colbourne et al. 2011). Functional genomic studies that target the identity of the regulators of plastic traits in both species will determine to what extent genetic mechanisms are species specific.
In the next section we review the results of earlier studies making use of the genome data when comparing aphid and Daphnia genes that may underlie phenotypic changes.
Early Functional Genetic Investigations of Phenotypic Plasticity in the Light of Genome Data
Caste Determination in Social Aphids—Implicating an Expanded Gene Family
By comparing mRNA profiles between soldier and normal morphs of the aphid Tuberaphis styraci, Kutsukake et al. (2004) revealed that one of the most up-regulated genes in the soldier morphs corresponded to a cysteine protease identified as a Cathepsin B. They demonstrated that this protease was produced within the gut of the soldiers and injected in the predators. This protein, once injected, acts as a toxin with insecticidal activity. Cathepsin B belongs to a multigenic family in insects, and two forms were identified in T. styraci: one specific to the soldier morph and one ubiquitously expressed in other morphs. The soldier-specific protein gene was shown to be under accelerated molecular evolution because of positive selection. This gene duplication and expression pattern was detected in other social aphid species, but the soldier specific form did not show accelerated evolution in these other lineages (Kutsukake et al. 2008). Close inspection of the pea aphid genome reveals a large scale expansion of the Cathepsin B genes with 28 copies (Rispe et al. 2008). There is substantial rate heterogeneity among gene copies with some evolving slowly and close to genes of other insects like Drosophila, and other copies evolving rapidly. There are clear indices for 16 paralogs of positive selection by the replacement of essential amino-acids within the protease active site. This observation suggests that these genes acquired new functions that remain to be discovered, in an aphid species (A. pisum) that do not develop soldier morphs.
Daphnia Plastic Response to Predators—Implicating Genes with Unknown Functions
Genome-wide tiling-path microarray experiments compared gene expression from second juvenile instars of D. pulex which were exposed and not exposed to dipteran predators (Chaoborus midges) (Colbourne et al. 2011). Significantly differentially regulated genes are found in classes of genes with, and without, protein homology. Examining the genomic regions with predicted gene models and at significant levels of differential expression, we detected 107 differentially regulated genes under the predator treatment that did not show significant differential regulation under other ecological treatments or life-history comparisons (metal toxicity, male/female). Of these candidates, 34 lack homology with proteins in other arthropod genomes. As a consequence, a significant proportion of the identified genes represent novel transcripts that require detailed functional characterization.
Wing Polyphenism in Aphids—Implicating Duplicated Genes with New Functions
Little (if anything) is known about the neuro-endocrine regulation of the wing polyphenism in aphids. Juvenile hormones are likely involved in the transduction events, but their role is still largely debated (Braendle et al. 2006). The genetic program responsible for wing development operates fully in winged morphs, but is interrupted at the larval stage of future unwinged morphs (Ishikawa et al. 2008). Alternative phenotypes are thus regulated at early developmental stages through repression in the wing development pathway. Not surprisingly, the principal genes involved in wing development described in Drosophila, such as apterous or decapentaplegic, were found to be duplicated in the pea aphid (Brisson et al. 2010). Interestingly, differential expression of these genes has been demonstrated between winged and unwinged individuals at different developmental stages reinforcing the observation from Daphnia that gene duplication and condition dependent diversification of gene regulation may play a role in the development of plastic phenotypes.
Reproductive Plasticity in Aphids—Implicating a Complex Gene Network
The alternation between asexual and sexual morphs occurs in autumn and is driven by the length of the photoperiod. More specifically, aphids sense the length of the night period (scotophase). This environmental input is necessary and sufficient for triggering a change in embryonic development that orientates towards a sexual phenotype producing gametes (males or females). Because of the telescoping of generations (embryos developing within the mothers), the adapted phenotype (sexual individuals) appears two or three generations after exposure to the new scotophase, and embryos can directly sense photoperiod change while inside the abdomen of the mother. The nature of the photoreceptors is not known but early transduction of the photoperiod signal is driven through neuro-secretory cells located in the dorsal part of the pars intercerebralis (the region between the two hemispheres of the brain) (Steel and Lees 1977). The nature of the neuro-secretory material released after photoperiod shortening is not known. Recently, based on the pea aphid genome and analysis of neuropeptides from the central nervous system, several hypotheses have been made for putative candidates responsible for neuro-regulation of photoperiod (Huybrechts et al. 2010) such as neuroparsin, insulins, eclosion hormone or GPA2/GPB5. Unpublished data from our group showed a localization of the SIFamide peptide in 4 cell bodies located in the same area that the neuro-secretory cells transducing the photoperiod signal (Verleyen and Tagu, unpublished). SIFamide is a neuropeptide ubiquitously found in insects (Nassel 2002) and is probably not the primarily responsible for photoperiod response, but could be involved in driving several inputs towards the thoracic ganglion mass or other endocrine organs that regulate hormone production (e.g., corpora allata). Transcriptomic approaches identified genetic programs in the head of aphids (containing central nervous system (CNS) structures) regulated by photoperiod shortening and reproductive switch (Le Trionnaire et al. 2007, 2009). Few changes were noted in transcript profiling of the grand-mothers of the sexual individuals, confirming early studies showing the absence of grand-maternal effect in reproductive polyphenism (Lees 1964). However, a large number (about 10% of the analysed transcripts) of genes were up- or down-regulated in heads of the mothers of the sexual individuals. The putative functions of several of these transcripts indicated the regulation of visual system, photoreception, neuro-endocrine signaling and regulation. For instance, two genes involved in the insulin pathway (a putative insulin receptor and a degrading insulin enzyme) were regulated such a way that a down regulation of the insulin pathway in aphids reared under long night might occur. In Drosophila, a decrease in the insulin pathway is correlated to a decrease in the JH pathway (Tu et al. 2005) and, as in aphids, it has been demonstrated that juvenile hormones regulate photoperiod signal transduction (Hardie et al. 1985). It is tempting to hypothesize that insulin could be an upstream regulator of juvenile hormones in short-day reared aphids. Annotation of the pea aphid genome identified an expansion of insulin genes (10 copies) (Huybrechts et al. 2010) and further molecular analyses are underway to check whether insulin genes are regulated during the switch of photoperiod. The transcriptomic approach also identified an unexpected genetic program highly regulated by photoperiod shortening; the cuticular proteins. These genes are among the most highly regulated, particularly those cuticular proteins that contain chitin-binding domains. This observation suggests that a softening of the cuticular network between cuticular proteins and chitin might occur in the heads of aphids reared under short-days (Le Trionnaire et al. 2009). In parallel, we noticed a down regulation of the pathway for the N-Beta alanine dopamine (NBAD), a metabolite that is stored in the cuticle and responsible for its sclerotization. The down regulation of the dopamine pathway suggests a role of dopamine due to photoperiod shortening (Gallot et al. 2010).
The targeted organs for photoperiod shortening are the ovaries, more specifically the germ cells for oocytes located in the germarium of the embryos of the future sexuals. In parthenogenetic embryos, these germ cells do not undergo meiosis and 2n oocyte-like cells directly enter embryogenesis in absence of male fertilization (Le Trionnaire et al. 2009). Annotation of the pea aphid genome for the genes involved in mitosis and meiosis showed some lineage-specific duplications of both mitotic regulators (e.g., Cdk1, Polo, Wee) and mitosis-related genes such as Smc6 (Srinivasan et al. 2010). It is intriguing to notice that these genes are single copies in other known insect genomes, and some are duplicated in the pea aphid and Daphnia (Schurko et al. 2009), both of which are capable of sexual and asexual reproduction.
Reproductive Plasticity in Daphnia—Implicating Meiosis Suppressor Genes
In Daphnia, sexual and asexual reproductive modes are ecologically relevant alternate strategies responsive to a variety of environmental cues. The genomic investigation of these alternative reproductive modes has focused on the annotation of genes involved with mitosis and meiosis and on the genetic underpinnings of the loss of the sexual reproductive mode. In an analysis of meiotic and mitotic genes in Daphnia, (Schurko et al. 2009) documented a pattern similar to aphids in lineage specific gene expansions. In particular, genes involved in cell cycle regulation and sister chromatid cohesion have expanded copy number compared to other arthropods. While no significant differences in regulation between sexual and parthenogenetic individuals were observed in this study, the expansion of meiotic gene families like Recq2 (a homologous recombination suppressor) may be associated with the ability to alternate between two reproductive modes. From an evolutionary perspective, recent work has identified a meiosis suppression factor responsible for the loss of the sexual reproductive mode in at least some populations (Lynch et al. 2008). Documenting the functional role of these expanded gene families in the switch to asexual reproduction in response to a cascade of cue detection and transduction remains a priority area of investigation. Since Daphnia are capable of switching from asexual to sexual reproduction and back to asexual within the lifetime of a single individual, a functional genomic approach will be useful to illuminate the transitions and pathways involved in alternate reproductive modes.
Environmental Sex Determination in Daphnia—Implicating a Juvenile Hormone Analog Pathway
Like for aphids, research in environmental sex determination mechanisms in Daphnia has implications for understanding invertebrate endocrine systems, genetic and epigenetic regulation of phenotypic plasticity, and crustacean developmental genetics in general, because JH and ecdysone pathways are linked (Mu and Leblanc 2004). Eads et al. (2007) reviewed this research, which stems from a discovery that sesquiterpenoid hormone methyl farnesoate (MF) stimulates the parthenogenetic production of males (Olmstead and Leblanc 2002). Molecular genetic studies show that sex in Daphnia is determined prior to the first embryonic cleavage (Olmstead and LeBlanc 2007) and that MF regulates numerous proteins including hemoglobin (Rider et al. 2005)—at least one of 7–8 genes arranged within a tandemly arranged cluster (Colbourne et al. 2011) containing an identifiable putative juvenoid response element (Gorr et al. 2006) — and a pair of vitellogenin genes that also contain a juvenoid response element in their shared promoter region (Tokishita et al. 2006). Access to the genome sequence enables a broader search of all annotated promoter regions for juvenoid response elements, which are indicative of potential targets for regulation, and for hormone receptors, whose identity serves to understand the extent of the upstream regulatory network controlling at least two significant plastic traits. So far, 25 putative nuclear receptors are annotated in the D. pulex genome sequence based on conserved DNA-binding domains (Thomson et al. 2009). An earlier study painstakingly revealed that the retinoid X receptor is differentially expressed in male and female Daphnia (Wang et al. 2007).
Gene expression experiments identified 55 differentially regulated genes when parthenogenetic females were exposed to MF, of which half had sequence homology to known proteins (Eads et al. 2007) that included hemoglobin, polysaccharide metabolism genes and chitin metabolism genes. The homologous gene sets underline a recurring association with ecdysone pathways. Continuing work in our laboratories make use of non-male producing isolates that are shown to be defective at different intervals along the MF signaling pathway (Rider et al. 2005).
Conclusion and Future Directions
We have shown here how ecologically relevant biological systems showing a wide range of continuous and discrete plastic traits could serve as models for studying the regulatory mechanisms underlying environmentally-induced developmental and physiological plasticity. Aphids and Daphnia share the feature of producing extremely diverse phenotypes from the same underlying genome. Ample genetic variation within populations for these plastic traits provides an opportunity to study the genomic underpinnings of plasticity and the ecological and evolutionary forces shaping these traits. We presented early progress in the understanding of the molecular, cellular and physiological mechanisms accounting for phenotypic plasticity in this duo, but recognize that these limited first steps contributing to the development of hypotheses that will form the basis of future investigations. We expect major advances in the field of developmental plasticity and Evo-Devo in aphids and Daphnia, enabled by the substantial efforts to gather genomic resources for each type of organism. The regulatory machinery of these two arthropods is currently being documented and analysed under ecologically relevant conditions leading to the production of alternative phenotypes. One intriguing feature emerging from comparative genomics of aphids and Daphnia is their common massive gene duplications. Gene duplication is a key mechanism for acquiring new functions since the original copy can be kept as a functional gene while the duplicated copy is free of selection pressure for sequence evolution and could acquire putative new functions (Innan and Kondrashov 2010). In Daphnia and aphids, gene duplication could represent a powerful evolutionary mechanism – through sequence variation and/or gene expression patterns - allowing enhanced genome functional flexibility necessary for exploring new adaptive traits.
Early studies on the control of polyphenic traits of aphids and Daphnia suggest a complexity of mechanisms involving a cascade of process from the reception of environmental stimuli triggering the development of alternate or modified phenotypes through gene and hormonal regulation. These modifications may involve regulation of micro RNAs and epigenetic effects of maternal induction. The regulatory mechanisms of phenotypic plasticity may not rely on a few “plasticity or robustness genes” but may rather involve complex gene networks organized in transcriptional modules (Ayroles et al. 2009). Investigating the role of epigenetics is one of the fastest growing fields in biological, evolutionary and ecological sciences. Epigenetic variation can be altered directly by the environment and has been shown to be responsible for some plastic traits (Bossdorf et al. 2008). The role of epigenetic process in controlling phenotypic traits can be now studied at a more detailed, functional level using QTL mapping or genome-wide approaches based on the detection of methylation marks on the DNA (Garfinkel et al. 2004). The generality of the discoveries in Daphnia and aphids remains an open ended question awaiting further investigation in other empirical systems. Arguably, the genome structure of Daphnia and aphids with their large gene content and high number of duplicate genes are unique among the fairly limited sample of arthropod genomes available. The number of additional genome sequences and systems with functional genomic tools will certainly increase dramatically in the next few years providing ample opportunities to validate the discoveries in Daphnia and aphids and generate additional hypotheses linking genome structure to phenotypic plasticity.
Finally, although theoretical models have suggested that polyphenism and polymorphism can evolve from each other (West-Eberhard 2003), how exactly the two are related evolutionarily or mechanistically and what are their respective costs and constraints remain unclear (Brisson et al. 2007). Gaining an understanding of the adaptive evolution of plasticity in these organisms will require linking genome structure with phenotype. Empirical investigation of the evolutionary and molecular bases of traits showing environmentally-induced plasticity (individual level) and genetic variation (population level), as defensive, reproductive or dispersal phenotypes in aphids and Daphnia, will help to clarify these issues and increase our understanding of why adaptive plasticity is not more widespread (Hilary et al. 2005).
Funding
This work was supported in part by the METACyt Initiative of Indiana University, funded through a major grant from the Lilly Endowment, Inc; and by the National Institutes of Health (grant number GM0782740). ANR Aphicibles, GnpAnnot and Holocentrisme funded in part research on the pea aphid. The scientific division of Plant health and Environment of the French Institute of Agriculture Research (INRA) is also acknowledged for its constant financial support to aphid researches.
Acknowledgments
We thank Prof. David Rand and the journal editors for the opportunity to contribute to this special issue on The Genetics and Genomics of Environmental Change. We also thank the numerous colleagues belonging to the aphid and Daphnia research communities that graciously shared many unpublished data on comparative and functional genomics of these 2 arthropods. Additional analyses were performed by wFleaBase, developed at the Genome Informatics Lab of Indiana University with support to Dr. Don Gilbert from the National Science Foundation and the National Institutes of Health. Coordination infrastructure for the Daphnia Genomics Consortium is provided by The Center for Genomics and Bioinformatics at Indiana University. Our work benefits from and contributes to the International Aphid Genomics and the Daphnia Genomics Consortia.
References
- Adamowicz SJ, Petrusek A, Colbourne JK, Hebert PDN, Witt JDS. The scale of divergence: a phylogenetic appraisal of intercontinental allopatric speciation in a passively dispersed freshwater zooplankton genus. Mol Phylogenet Evol. 2009;50:423–436. doi: 10.1016/j.ympev.2008.11.026. [DOI] [PubMed] [Google Scholar]
- Agrawal AA, Laforsch C, Tollrian R. Transgenerational induction of defences in animals and plants. Nature. 1999;401:60–63. [Google Scholar]
- Alekseev V, Lampert W. Maternal control of resting-egg production in Daphnia. Nature. 2001;414:899–901. doi: 10.1038/414899a. [DOI] [PubMed] [Google Scholar]
- Aubin-Horth N, Renn SCP. Genomic reaction norms: using integrative biology to understand molecular mechanisms of phenotypic plasticity. Mol Ecol. 2009;18:3763–3780. doi: 10.1111/j.1365-294X.2009.04313.x. [DOI] [PubMed] [Google Scholar]
- Ayroles JF, Carbone MA, Stone EA, Jordan KW, Lyman RF, Magwire MM, Rollmann SM, Duncan LH, Lawrence F, Anholt RRH, et al. Systems genetics of complex traits in Drosophila melanogaster. Nat Genet. 2009;41:299–307. doi: 10.1038/ng.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin JM. A new factor in evolution. Am Nat. 1896;30:441–451. [Google Scholar]
- Banta AM, Brown LA. Control of male and sexual egg production. In: Banta AM, editor. Studies on the physiology, genetics and evoltuion of some Cladocera. Washington (DC): Carnegie Institute; 1939. pp. 106–129. [Google Scholar]
- Barry MJ. Progress toward understanding the neurophysiological basis of predator-induced morphology in Daphnia pulex. Physiol Biochem Zool. 2002;75:179–186. doi: 10.1086/339389. [DOI] [PubMed] [Google Scholar]
- Beaton MJ, Hebert PDN. The cellular basis of divergent head morphologies in Daphnia. Limnol Oceanogr. 1997;42:346–356. [Google Scholar]
- Behura SK. Insect microRNAs: structure, function and evolution. Insect Biochem Mol Biol. 2007;37:3–9. doi: 10.1016/j.ibmb.2006.10.006. [DOI] [PubMed] [Google Scholar]
- Berg K. Cyclic reproduction, sex determination and depression in the Cladocera. Biol Rev. 1934;9:139–174. [Google Scholar]
- Berrigan D, Scheiner SM. Modeling the evolution of phenotypic plasticity. In: DeWitt TJ, Scheiner SM, editors. Phenotypic plasticity: functional and conceptual advances. New York: Oxford University Press; 2004. [Google Scholar]
- Bossdorf O, Richards CL, Pigliucci M. Epigenetics for ecologists. Ecol Lett. 2008;11:106–115. doi: 10.1111/j.1461-0248.2007.01130.x. [DOI] [PubMed] [Google Scholar]
- Braendle C, Davis GK, Brisson JA, Stern DL. Wing dimorphism in aphids. Heredity. 2006;97:192–199. doi: 10.1038/sj.hdy.6800863. [DOI] [PubMed] [Google Scholar]
- Braendle C, Friebe I, Caillaud MC, Stern DL. Genetic variation for an aphid wing polyphenism is genetically linked to a naturally occurring wing polymorphism. Proc R Soc B Biol Sci. 2005;272:657–664. doi: 10.1098/rspb.2004.2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braendle C, Miura T, Bickel R, Shingleton AW, Kambhampati S, Stern DL. Developmental origin and evolution of bacteriocytes in the aphid- Buchnera symbiosis. PloS Biol. 2003;1:70–76. doi: 10.1371/journal.pbio.0000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brisson JA. Aphid wing dimorphisms: linking environmental and genetic control of trait variation. Philos Trans R Soc B Biol Sci. 2010;365:605–616. doi: 10.1098/rstb.2009.0255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brisson JA, Davis GK, Stern DL. Common genome-wide patterns of transcript accumulation underlying the wing polyphenism and polymorphism in the pea aphid (Acyrthosiphon pisum) Evol Dev. 2007;9:338–346. doi: 10.1111/j.1525-142X.2007.00170.x. [DOI] [PubMed] [Google Scholar]
- Brisson JA, Ishikawa A, Miura T. Wing development genes of the pea aphid and differential gene expression between winged and unwinged morphs. Insect Mol Biol. 2010;19:63–73. doi: 10.1111/j.1365-2583.2009.00935.x. [DOI] [PubMed] [Google Scholar]
- Brooks DR, McLennan DA. Phylogeny, ecology, and behaviour: a research program in comparative biology. 1991;i–xii [Google Scholar]
- Callahan HS, Dhanoolal N, Ungerer MC. Plasticity genes and plasticity costs: a new approach using an Arabidopsis recombinant inbred population. New Phytol. 2005;166:129–139. doi: 10.1111/j.1469-8137.2005.01368.x. [DOI] [PubMed] [Google Scholar]
- Carlborg O, Haley CS. Epistasis: too often neglected in complex trait studies? Nat Rev Genet. 2004;5:618–U614. doi: 10.1038/nrg1407. [DOI] [PubMed] [Google Scholar]
- Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho GR, Hughes RN. The effect of food availability, female culture-density and photoperiod on ephippia production in Daphnia magna straus (Crustacea, Cladocera) Freshw Biol. 1983;13:37–46. [Google Scholar]
- Charmantier A, McCleery RH, Cole LR, Perrins C, Kruuk LEB, Sheldon BC. Adaptive phenotypic plasticity in response to climate change in a wild bird population. Science. 2008;320:800–803. doi: 10.1126/science.1157174. [DOI] [PubMed] [Google Scholar]
- Chevin L-M, Lande R, Mace GM. Adaptation, plasticity, and extinction in a changing environment: towards a predictive theory. PLoS Biol. 2010;8:e1000357. doi: 10.1371/journal.pbio.1000357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colbourne JK, Hebert PDN. The systematics of North American Daphnia (Crustacea: Anomopoda): a molecular phylogenetic approach. Philos Trans R Soc Lond Ser B Biol Sci. 1996;351:349–360. doi: 10.1098/rstb.1996.0028. [DOI] [PubMed] [Google Scholar]
- Colbourne JK, Hebert PDN, Taylor DJ. Evolutionary origins of phenotypic diversity in Daphnia. In: Givnish TJ, Sytsma KJ, editors. Molecular evolution and adaptive radiation. Cambridge University Press; 1997. pp. 163–188. [Google Scholar]
- Colbourne JK, Pfrender ME, Gilbert D, Thomas WK, Tucker A, Oakley TH, Tokishita S, Aerts A, Arnold GJ, Kumar Basu M, et al. The ecoresponsive genome of Daphnia pulex. Science. 2011;331:555–561. doi: 10.1126/science.1197761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colbourne JK, Singan VR, Gilbert DG. wFleaBase: the Daphnia genome database. BMC Bioinformatics. 2005;6:45. doi: 10.1186/1471-2105-6-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cousyn C, De Meester L, Colbourne JK, Brendonck L, Verschuren D, Volckaert F. Rapid, local adaptation of zooplankton behavior to changes in predation pressure in the absence of neutral genetic changes. Proc Natl Acad Sci U S A. 2001;98:6256–6260. doi: 10.1073/pnas.111606798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison J. Activation of ephippial egg of Daphnia pulex. J Gen Physiol. 1969;53:562–575. doi: 10.1085/jgp.53.5.562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Meester L, Dejager H. Hatching of Daphnia sexual eggs .1. Intraspecific differences in the hatching responses of Daphnia magna eggs. Freshw Biol. 1993;30:219–226. [Google Scholar]
- De Meester L, Gomez A, Simon J-C, Moya A, Font E. Evolution: from molecules to ecosystems. 2004. Evolutionary and ecological genetics of cyclical parthenogens; pp. 122–134. [Google Scholar]
- Deng HW. Environmental and genetic control of sexual reproduction in Daphnia. Heredity. 1996;76:449–458. [Google Scholar]
- Dixon AFG. Aphid ecology: an optimization approach Aphid ecology: an optimization approach. (2nd ed.) 1998;i–ix:1–300. [Google Scholar]
- Eads BD, Andrews J, Colbourne JK. Ecological genomics in Daphnia: stress responses and environmental sex determination. Heredity. 2007;100:184–190. doi: 10.1038/sj.hdy.6800999. [DOI] [PubMed] [Google Scholar]
- Ellegren H, Parsch J. The evolution of sex-biased genes and sex-biased gene expression. Nat Rev Genet. 2007;8:689–698. doi: 10.1038/nrg2167. [DOI] [PubMed] [Google Scholar]
- Fisk DL, Latta LCIV, Knapp RA, Pfrender ME. Rapid evolution in response to introduced predators I: rates and patterns of morphological and life-history trait divergence. BMC Evol Biol. 2007;7:1–11. doi: 10.1186/1471-2148-7-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint J, Mackay TFC. Genetic architecture of quantitative traits in mice, flies, and humans. Genome Res. 2009;19:723–733. doi: 10.1101/gr.086660.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel W, Luttbeg B, Sih A, Tollrian R. Environmental tolerance, heterogeneity, and the evolution of reversible plastic responses. Am Nat. 2005;166:339–353. doi: 10.1086/432558. [DOI] [PubMed] [Google Scholar]
- Gallot A, Rispe C, Leterme N, Gauthier J-P, Jaubert-Possamai S, Tagu D. Cuticular proteins and seasonal photoperiodism in aphids. Insect Biochem Mol Biol. 2010;40:235–240. doi: 10.1016/j.ibmb.2009.12.001. [DOI] [PubMed] [Google Scholar]
- Garfinkel MD, Sollars VE, Lu X, Ruden DM. Multigenerational selection and detection of altered histone acetylation and methylation patterns: toward a quantitative epigenetics in Drosophila. Methods Mol Biol. 2004;287:151–168. doi: 10.1385/1-59259-828-5:151. [DOI] [PubMed] [Google Scholar]
- Gerlach D, Kriventseva EV, Rahman N, Vejnar CE, Zdobnov EM. miROrtho: computational survey of microRNA genes. Nucleic Acids Res. 2009;37:D111–D117. doi: 10.1093/nar/gkn707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert D. OrthoMCL clustering among 13 arthropod proteomes. 2008. Available from: http://insects.eugenes.org/arthropods/ [Google Scholar]
- Gorr TA, Rider CV, Wang HY, Olmstead AW, LeBlanc GA. A candidate juvenoid hormone receptor cis-element in the Daphnia magna hb2 hemoglobin gene promoter, Mol Cell Endocrinol. 2006;247:91–102. doi: 10.1016/j.mce.2005.11.022. [DOI] [PubMed] [Google Scholar]
- Gutteling EW, Riksen JAG, Bakker J, Kammenga JE. Mapping phenotypic plasticity and genotype–environment interactions affecting life-history traits in Caenorhabditis elegans. Heredity. 2007;98:28–37. doi: 10.1038/sj.hdy.6800894. [DOI] [PubMed] [Google Scholar]
- Hales DF, Mittler TE. Chromosomal sex determination in aphids controlled by juvenile-hormone. Genome. 1987;29:107–109. [Google Scholar]
- Hanazato T. Pesticides as chemical-agents inducing helmet formation in Daphnia ambigua. Freshw Biol. 1991;26:419–424. [Google Scholar]
- Hanazato T. Insecticide inducing helmet development in Daphnia ambigua. Arch Hydrobiol. 1992;123:451–457. [Google Scholar]
- Hardie J, Lees AD, Kerkut GA, Gilbert LI. Endocrine control of polymorphism and polyphenism. Comprehensive insect physiology, biochemistry and pharmacology. Volume 8. Endocrinology. 1985;2:441–490. [Google Scholar]
- Hilary S, Callahan ND, Ungerer MC. Plasticity genes and plasticity costs: a new approach using an Arabidopsis recombinant inbred population. New Phytol. 2005;166:129–140. doi: 10.1111/j.1469-8137.2005.01368.x. [DOI] [PubMed] [Google Scholar]
- Hurst LD, Williams EJB, Pal C. Natural selection promotes the conservation of linkage of co-expressed genes. Trends Genet. 2002;18:604–606. doi: 10.1016/s0168-9525(02)02813-5. [DOI] [PubMed] [Google Scholar]
- Huybrechts J, Bonhomme J, Minoli S, Prunier-Leterme N, Dombrovsky A, Abdel-Latief M, Robichon A, Veenstra JA, Tagu D. Neuropeptide and neurohormone precursors in the pea aphid Acyrthosiphon pisum. Insect Mol Biol. 2010;19:87–95. doi: 10.1111/j.1365-2583.2009.00951.x. [DOI] [PubMed] [Google Scholar]
- Innan H, Kondrashov F. The evolution of gene duplications: classifying and distinguishing between models. Nat Rev Genet. 2010;11:97–108. doi: 10.1038/nrg2689. [DOI] [PubMed] [Google Scholar]
- Innes DJ, Dunbrack RL. Sex allocation variation in Daphnia pulex. J Evol Biol. 1993;6:559–575. [Google Scholar]
- Innes DJ, Singleton DR. Variation in allocation to sexual and asexual reproduction among clones of cyclically parthenogenetic Daphnia pulex (Crustacea: Cladocera) Biol J Linn Soc. 2000;71:771–787. [Google Scholar]
- Ishikawa A, Miura T. Morphological differences between wing morphs of two Macrosiphini aphid species, Acyrthosiphon pisum and Megoura crassicauda (Hemiptera, Aphididae) Sociobiology. 2007;50:881–893. [Google Scholar]
- Ishikawa A, Hongo S, Miura T. Morphological and histological examination of polyphonic wing formation in the pea aphid Acyrthosiphon pisum (Hemiptera, Hexapoda) Zoomorphology. 2008;127:121–133. [Google Scholar]
- Johannes F, Porcher E, Teixeira FK, Saliba-Colombani V, Simon M, Agier N, Bulski A, Albuisson J, Heredia F, Audigier P, et al. Assessing the impact of transgenerational epigenetic variation on complex traits. PLoS Genet. 2009;5:e1000530. doi: 10.1371/journal.pgen.1000530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juenger TE, Sen S, Stowe KA, Simms EL. Epistasis and genotype-environment interaction for quantitative trait loci affecting flowering time in Arabidopsis thaliana. Genetica. 2005;123:87–105. doi: 10.1007/s10709-003-2717-1. [DOI] [PubMed] [Google Scholar]
- Kunert G, Otto S, Rose USR, Gershenzon J, Weisser WW. Alarm pheromone mediates production of winged dispersal morphs in aphids. Ecol Lett. 2005;8:596–603. [Google Scholar]
- Kutsukake M, Nikoh N, Shibao H, Rispe C, Simon JC, Fukatsu T. Evolution of soldier-specific venomous protease in social aphids. Mol Biol Evol. 2008;25:2627–2641. doi: 10.1093/molbev/msn203. [DOI] [PubMed] [Google Scholar]
- Kutsukake M, Shibao H, Nikoh N, Morioka M, Tamura T, Hoshino T, Ohgiya S, Fukatsu T. Venomous protease of aphid soldier for colony defense. Proc Natl Acad Sci U S A. 2004;101:11338–11343. doi: 10.1073/pnas.0402462101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laforsch C, Tollrian R. Embryological aspects of inducible morphological defenses in Daphnia. J Morphol. 2004;262:701–707. doi: 10.1002/jmor.10270. [DOI] [PubMed] [Google Scholar]
- Latta LCIV, Bakelar JW, Knapp RA, Pfrender ME. Rapid evolution in response to introduced predators II: the contribution of adaptive plasticity. BMC Evol Biol. 2007;7:1–9. doi: 10.1186/1471-2148-7-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Trionnaire G, Francis F, Jaubert S, Bonhomme J, De Pauw E, Gauthier J-P, Haubruge E, Legeai F, Prunier-Leterme N, Simon J-C, et al. Transcriptomic and proteomic analyses of seasonal photoperiodism in the pea aphid. BMC Genomics. 2009;10(456):14. doi: 10.1186/1471-2164-10-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Trionnaire G, Hardie J, Jaubert-Possamai S, Simon JC, Tagu D. Shifting from clonal to sexual reproduction in aphids: physiological and developmental aspects. Biol Cell. 2008;100:441–451. doi: 10.1042/BC20070135. [DOI] [PubMed] [Google Scholar]
- Le Trionnaire G, Jaubert S, Sabater-Munoz B, Benedetto A, Bonhomme J, Prunier-Leterme N, Martinez-Torres D, Simon JC, Tagu D. Seasonal photoperiodism regulates the expression of cuticular and signalling protein genes in the pea aphid. Insect Biochem Mol Biol. 2007;37:1094–1102. doi: 10.1016/j.ibmb.2007.06.008. [DOI] [PubMed] [Google Scholar]
- Lees AD. Location of photoperiodic receptors in aphid Megoura viciae buckton. J Exp Biol. 1964;41:119. doi: 10.1242/jeb.41.1.119. [DOI] [PubMed] [Google Scholar]
- Legeai F, Rizk G, Walsh T, Edwards O, Gordon K, Lavenier D, Leterme N, Méreau A, Nicolas J, Tagu D, et al. Bioinformatic prediction, deep sequencing of microRNAs and their role in phenotypic plasticity in the pea aphid, Acyrthosiphon pisum. BMC Genomics. 2010;11:281. doi: 10.1186/1471-2164-11-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legeai F, Shigenobu S, Gauthier J-P, Colbourne JK, Rispe C, Collin O, Richards S, Wilson ACC, Tagu D. AphidBase: a centralized bioinformatic resource for annotation of the pea aphid genome. Insect Mol Biol. 2010;19:5–12. doi: 10.1111/j.1365-2583.2009.00930.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Seyfert A, Eads B, Williams E. Localization of the genetic determinants of meiosis suppression in Daphnia pulex. Genetics. 2008;180:317–327. doi: 10.1534/genetics.107.084657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miner BG, Sultan SE, Morgan SG, Padilla DK, Relyea RA. Ecological consequences of phenotypic plasticity. Trends Ecol Evol. 2005;20:685–692. doi: 10.1016/j.tree.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Mitchell-Olds T, Schmitt J. Genetic mechanisms and evolutionary significance of natural variation in Arabidopsis. Nature. 2006;441:947–952. doi: 10.1038/nature04878. [DOI] [PubMed] [Google Scholar]
- Miura T, Braendle C, Shingleton A, Sisk G, Kambhampati S, Stern DL. A comparison of parthenogenetic and sexual embryogenesis of the pea aphid Acyrthosiphon pisum (Hemiptera: Aphidoidea) J Exp Zool Part B Mol Develop Evol. 2003;295B:59–81. doi: 10.1002/jez.b.3. [DOI] [PubMed] [Google Scholar]
- Mu XY, Leblanc GA. Cross communication between signaling pathways: juvenoid hormones modulate ecdysteroid activity in a crustacean. J Exp Zool Part A Comp Exp Biol. 2004;301A:793–801. doi: 10.1002/jez.a.104. [DOI] [PubMed] [Google Scholar]
- Nassel DR. Neuropeptides in the nervous system of Drosophila and other insects: multiple roles as neuromodulators and neurohormones. Progress Neurobiol. 2002;68:1–84. doi: 10.1016/s0301-0082(02)00057-6. [DOI] [PubMed] [Google Scholar]
- Nijhout HF. Control mechanisms of polyphenic development in insects—in polyphenic development, environmental factors alter same aspects of development in an orderly and predictable way. Bioscience. 1999;49:181–192. [Google Scholar]
- Ollivier M, Legeai F, Rispe C. Comparative analysis of the Acyrthosiphon pisum genome and expressed sequence tag-based gene sets from other aphid species. Insect Mol Biol. 2010;19:33–45. doi: 10.1111/j.1365-2583.2009.00976.x. [DOI] [PubMed] [Google Scholar]
- Olmstead AW, Leblanc GA. Juvenoid hormone methyl farnesoate is a sex determinant in the crustacean Daphnia magna. J Exp Zool. 2002;293:736–739. doi: 10.1002/jez.10162. [DOI] [PubMed] [Google Scholar]
- Olmstead AW, LeBlanc GA. Insecticidal juvenile hormone analogs stimulate the production of male offspring in the crustacean Daphnia magna. Environ Health Perspect. 2003;111:919–924. doi: 10.1289/ehp.5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmstead AW, LeBlanc GA. The environmental-endocrine basis of gynandromorphism (intersex) in a crustacean. Int J Biol Sci. 2007;3:77–84. doi: 10.7150/ijbs.3.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi M, Nuttall R, Edwards P, Minor J, Naiman D, Lu JN, Doctolero M, Vainer M, Chan C, Malley J, et al. A survey of ovary-, testis-, and soma-biased gene expression in Drosophila melanogaster adults. Genome Biol. 2004;5(6):R40. doi: 10.1186/gb-2004-5-6-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrusek A, Tollrian R, Schwenk K, Haas A, Laforsch C. A "crown of thorns' is an inducible defense that protects Daphnia against an ancient predator. Proc Natl Acad Sci USA. 2009;106:2248–2252. doi: 10.1073/pnas.0808075106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfrender ME, Deng H-W. Environmental and genetic control of diapause termination in Daphnia. Adv Limnol. 1998;52:237–251. [Google Scholar]
- Pfrender ME, Spitze K, Lehman N. Multi-locus genetic evidence for rapid ecologically based speciation in Daphnia. Mol Ecol. 2000;9:1717–1735. doi: 10.1046/j.1365-294x.2000.01062.x. [DOI] [PubMed] [Google Scholar]
- Pigliucci M. Evolution of phenotypic plasticity: where are we going now? Trends Ecol Evol. 2005;20:481–486. doi: 10.1016/j.tree.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Pohnert G, Steinke M, Tollrian R. Chemical cues, defence metabolites and the shaping of pelagic interspecific interactions. Trends Ecol Evol. 2007;22:198–204. doi: 10.1016/j.tree.2007.01.005. [DOI] [PubMed] [Google Scholar]
- Rice SH. A general population genetic theory for the evolution of developmental interactions. Proc Natl Acad Sci U S A. 2002;99:15518–15523. doi: 10.1073/pnas.202620999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice SH. Developmental associations between traits: covariance and beyond. Genetics. 2004;166:513–526. doi: 10.1534/genetics.166.1.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rider CV, Gorr TA, Olmstead AW, Wasilak BA, Leblanc GA. Stress signaling: coregulation of hemoglobin and male sex determination through a terpenoid signaling pathway in a crustacean. J Exp Biol. 2005;208:15–23. doi: 10.1242/jeb.01343. [DOI] [PubMed] [Google Scholar]
- Rispe C, Kutsukake M, Doublet V, Hudaverdian S, Legeai F, Simon JC, Tagu D, Fukatsu T. Large gene family expansion and variable selective pressures for cathepsin B in aphids. Mol Biol Evol. 2008;25:5–17. doi: 10.1093/molbev/msm222. [DOI] [PubMed] [Google Scholar]
- Sabater-Munoz B, Legeai F, Rispe C, Bonhomme J, Dearden P, Dossat C, Duclert A, Gauthier JP, Ducray DG, Hunter W, et al. Large-scale gene discovery in the pea aphid Acyrthosiphon pisum (Hemiptera) Genome Biol. 2006;7(3):R21. doi: 10.1186/gb-2006-7-3-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S. From Reaktionsnorm to the evolution of adaptive plasticity: a historical sketch, 1909–1999. In: DeWitt TJ, Scheiner SM, editors. Phenotypic plasticity: functional and conceptual advances. New York: Oxford University Press; 2004. [Google Scholar]
- Scheiner SM. Genetics and evolution of phenotypic plasticity. Ann Rev Ecol Syst. 1993;24:35–68. [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Scholkopf B, Weigel D, Lohmann JU. A gene expression map of Arabidopsis thaliana development. Nat Genet. 2005;37:501–506. doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
- Schurko AM, Logsdon JM, Eads BD. Meiosis genes in Daphnia pulex and the role of parthenogenesis in genome evolution. BMC Evol Biol. 2009;9:78. doi: 10.1186/1471-2148-9-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scoville AG, Pfrender ME. Phenotypic plasticity facilitates recurrent rapid adaptation to introduced predators. Proc Natl Acad Sci. 2010;107:4260–4263. doi: 10.1073/pnas.0912748107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sempere LF, Dubrovsky EB, Dubrovskaya VA, Berger EM, Ambros V, Ros A. The expression of the let-7 small regulatory RNA is controlled by ecdysone duriniz metamorphosis in Drosophila melanogaster. Dev Biol. 2002;244:170–179. doi: 10.1006/dbio.2002.0594. [DOI] [PubMed] [Google Scholar]
- Simon JC, Baumann S, Sunnucks P, Hebert PDN, Pierre JS, Le Gallic JF, Dedryver CA. Reproductive mode and population genetic structure of the cereal aphid Sitobion avenae studied using phenotypic and microsatellite markers. Mol Ecol. 1999;8:531–545. doi: 10.1046/j.1365-294x.1999.00583.x. [DOI] [PubMed] [Google Scholar]
- Simon JC, Leterme N, Latorre A. Molecular markers linked to breeding system differences in segregating and natural populations of the cereal aphid Rhopalosiphum padi L. Mol Ecol. 1999;8:965–973. doi: 10.1046/j.1365-294x.1999.00648.x. [DOI] [PubMed] [Google Scholar]
- Simon JC, Rispe C, Sunnucks P. Ecology and evolution of sex in aphids. Trends Ecol Evol. 2002;17:34–39. [Google Scholar]
- Srinivasan DG, Fenton B, Jaubert-Possamai S, Jaouannet M. Analysis of meiosis and cell cycle genes of the facultatively asexual pea aphid, Acyrthosiphon pisum (Hemiptera: Aphididae) Insect Mol Biol. 2010;19:229–239. doi: 10.1111/j.1365-2583.2009.00960.x. [DOI] [PubMed] [Google Scholar]
- Steel CGH, Lees AD. Role of neurosecretion in photoperiodic control of polymorphism in aphid Megoura viciae. J Exp Biol. 1977;67:117. doi: 10.1242/jeb.67.1.117. [DOI] [PubMed] [Google Scholar]
- Stern DL, Foster WA. The evolution of soldiers in aphids. Biol Rev Camb Philos Soc. 1996;71:27–79. doi: 10.1111/j.1469-185x.1996.tb00741.x. [DOI] [PubMed] [Google Scholar]
- Stibor H, Luning J. Predator-induced phenotypic variation in the pattern of growth and reproduction in Daphnia hyalina (Crustacea, Cladocera) Funct Ecol. 1994;8:97–101. [Google Scholar]
- Stross RG, Hill JC. Diapause induction in Daphnia requires 2 stimuli. Science. 1965;150:1462. doi: 10.1126/science.150.3702.1462. [DOI] [PubMed] [Google Scholar]
- Tagu D, Klingler JP, Moya A, Simon J-C. Early progress in aphid genomics and consequences for plant–aphid interactions studies. Mol Plant Microbe Interact. 2008;21:701–708. doi: 10.1094/MPMI-21-6-0701. [DOI] [PubMed] [Google Scholar]
- Tagu D, Sabater-Munoz B, Simon JC. Deciphering reproductive polyphenism in aphids. Invertebr Reprod Dev. 2005;48:71–80. [Google Scholar]
- The International Aphid Genomics Consortium. Genome sequence of the pea aphid Acyrthosiphon pisum. PLoS Biol. 2010;8:e1000313. doi: 10.1371/journal.pbio.1000313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson SA, Wang YH, LeBlanc GA. Inventory of nuclear hormone receptors of Daphnia pulex: insights into function and evolution. BMC Genomics. 2009;10:500. doi: 10.1186/1471-2164-10-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokishita S, Kato Y, Kobayashi T, Nakamura S, Ohta T, Yamagata H. Organization and repression by juvenile hormone of a vitellogenin gene cluster in the crustacean, Daphnia magna. Biochem Biophys Res Commun. 2006;345:362–370. doi: 10.1016/j.bbrc.2006.04.102. [DOI] [PubMed] [Google Scholar]
- Tollrian R. Predator induced defenses: Costs, life-history shifts and maternal effects in Daphnia pulex. Ecology. 1995;76:1691–1705. [Google Scholar]
- Tollrian R, Dodson SI. Predator induced defenses in cladocerans. In: Tollrian R, Harvell CD, editors. The ecology and evolution of inducible defenses. Princeton (NJ): Princeton University Press; 1999. [Google Scholar]
- Tollrian R, Harvell CD, editors. The Ecology and evolution of inducible defenses. Princeton (NJ): Princeton University Press; 1999. [Google Scholar]
- Tollrian R, Leese F. Ecological genomics: steps towards unraveling the genetic basis of inducible defenses in Daphnia. BMC Biol. 2010;8:51. doi: 10.1186/1741-7007-8-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu MP, Yin CM, Tatar M. Mutations in insulin signaling pathway alter juvenile hormone synthesis in Drosophila melanogaster. Gen Comp Endocrinol. 2005;142:347–356. doi: 10.1016/j.ygcen.2005.02.009. [DOI] [PubMed] [Google Scholar]
- Ungerer MC, Halldorsdottir SS, Purugganan MA, Mackay TFC. Genotype-environment interactions at quantitative trait loci affecting inflorescence development in Arabidopsis thaliana. Genetics. 2003;165:353–365. doi: 10.1093/genetics/165.1.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Via S, Gomulkiewicz R, Dejong G, Scheiner SM, Schlichting CD, Vantienderen PH. Adaptive phenotypic plasticity—consensus and controversy. Trends Ecol Evol. 1995;10:212–217. doi: 10.1016/s0169-5347(00)89061-8. [DOI] [PubMed] [Google Scholar]
- Walsh TK, Brisson JA, Robertson HM, Gordon K, Jaubert-Possamai S, Tagu D, Edwards OR. A functional DNA methylation system in the pea aphid Acyrthosiphon pisum. Insect Mol Biol. 2010;19:215–228. doi: 10.1111/j.1365-2583.2009.00974.x. [DOI] [PubMed] [Google Scholar]
- Wang YH, Wang GR, LeBlanc GA. Cloning and characterization of the retinoid X receptor from a primitive crustacean Daphnia magna. Gen Comp Endocrinol. 2007;150:309–318. doi: 10.1016/j.ygcen.2006.08.002. [DOI] [PubMed] [Google Scholar]
- Weinig C, Schmitt J. Environmental effects on the expression of quantitative trait loci and implications for phenotypic evolution. Bioscience. 2004;54:627–635. [Google Scholar]
- Honeybee Genome Sequencing Consortium Insights into social insects from the genome of the honeybee Apis mellifera. Nature. 2006;443:931–949. doi: 10.1038/nature05260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werren JH, Richards S, Desjardins CA, Niehuis O, Gadau J, Colbourne JK, Beukeboom LW, Desplan C, Elsik CG, Grimmelikhuijzen CJP, et al. Insights from three genomes of Nasonia: insect natural enemies and emerging genetic model. Science. 2010;327:343–348. [Google Scholar]
- West-Eberhard MJ. Developmental plasticity and evolution. Developmental plasticity and evolution. Oxford University Press; 2003. 794 pp. [Google Scholar]
- Williams EJB, Bowles DJ. Coexpression of neighboring genes in the genome of Arabidopsis thaliana. Genome Res. 2004;14:1060–1067. doi: 10.1101/gr.2131104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf JB, Brodie ED, III, Wade MJ. The genotype–environment and evolution when the environment contains genes. In: DeWitt TJ, Scheiner SM, editors. Phenotypic plasticity: functional and conceptual advances. New York: Oxford Univ. Press; 2004. [Google Scholar]
- Woltereck R. Weitere experimentelle Untersuchungen uber Art veranderung, speziell uber das Wesen quantitativer Artunterschiede bei Daphniden. Verh D zool GesLeipzig. 1909;19:110–173. [Google Scholar]
- Yampolsky LY. Reaction norms of quantitative traits and genotype–environment interactions in Daphnia. Genetika. 1992;28:85–92. [Google Scholar]
- Zhang Y, Sturgill D, Parisi M, Kumar S, Oliver B. Constraint and turnover in sex-biased gene expression in the genus Drosophila. Nature. 2007;450:233–237. doi: 10.1038/nature06323. [DOI] [PMC free article] [PubMed] [Google Scholar]