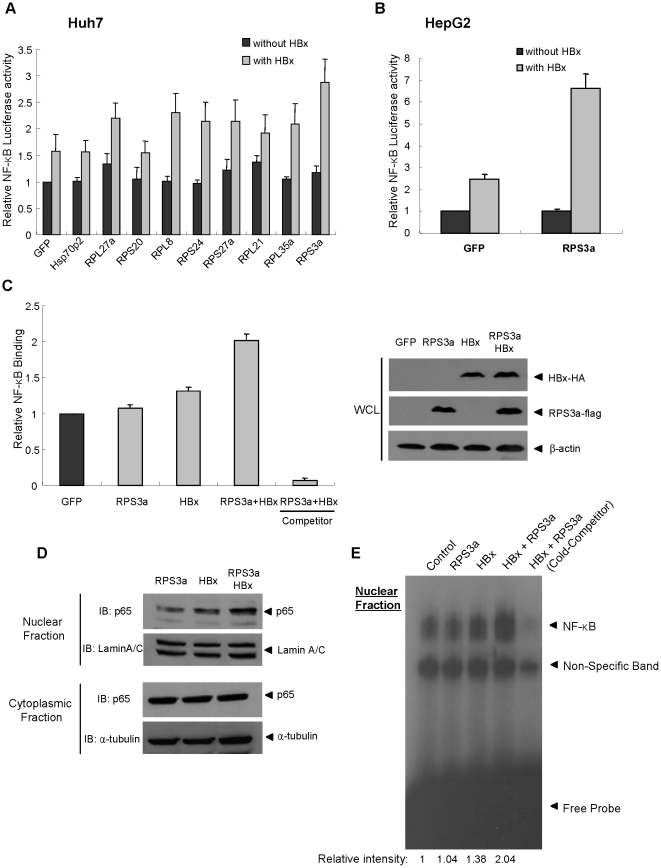
Figure 1. RPS3a enhances the HBx-induced NF-κB Signaling.
(A–B) Relative NF-κB activity in Huh7 and HepG2 liver cells after co-transfection of pNF-κB-Luc(0.25 µg), pEGFP(0.4 µg) (control plasmid) and the indicated plasmid(0.4 µg) with/without pEG-HBx(0.4 µg). (C) ELISA of NF-κB activation in nucleus. Using nuclear extracts (4 µg), the binding between activated NF-κB subunit in nucleus and plate-bound NF-κB consensus DNA oligomer was measured by chemiluminescence (left). Free unbound NF-κB consensus DNA oligomer was used as a competitor. Total expression levels of HBx and RPS3a in whole cell lysate (WCL) were determined by indicated antibodies (right). (D) Western blot analysis of p65 translocation in Huh7 cells. Lamin A/C and α-tubulin were used for loading controls of nuclear and cytoplasmic fractions, respectively. The loading amount in cytoplasmic fractions is 5-fold dilution than that of the nuclear fractions. (E) Electrophoretic Mobility Shift Assay (EMSA). Separated nuclear extracts (3 µg) were subjected to in vitro binding with [P32]-labeled NF-κB consensus DNA oligomer probe. pEGFP vector was used as control vector. As a cold competitor, excess (30-fold) non-labeled NF-κB consensus oligomer was used. Relative binding intensity was calculated by using densitometry.