Figure 6. Summary of experimental and analytical approaches used, and significant findings.
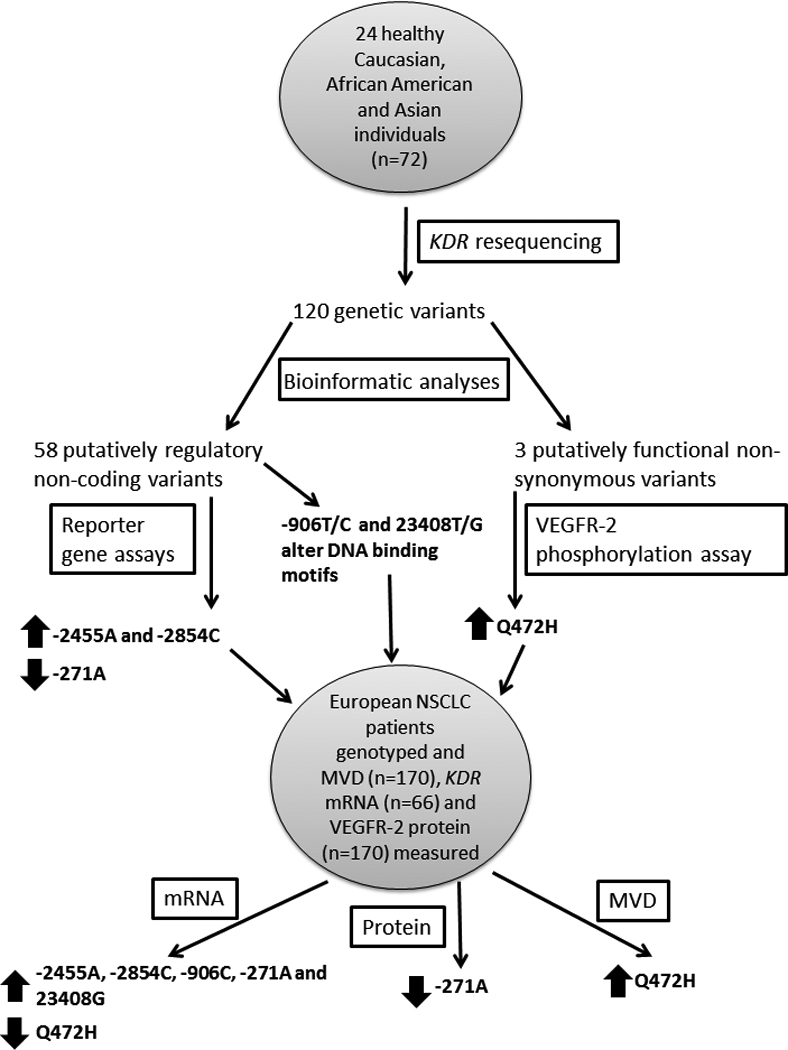
KDR was resequenced in three ethnic groups, each containing 24 healthy individuals. The genetic variants identified from the resequencing were analyzed for function using bioinformatic tools. Putatively functional SNPs were examined using reporter gene or VEGFR-2 phosphorylation assays. Functional SNPs, including two further putatively functional SNPs, were genotyped in a cohort of European NSCLC patients. These genotypes were tested for associations with KDR mRNA, VEGFR-2 protein and MVD levels from matching tumor samples. SNPs significantly (p<0.05) associated with a phenotype of interest are designated by a block arrow. An up arrow indicates that the variant is associated with increased levels of the phenotype; conversely, a down arrow indicates that the variant is associated with decreased levels of the phenotype.
