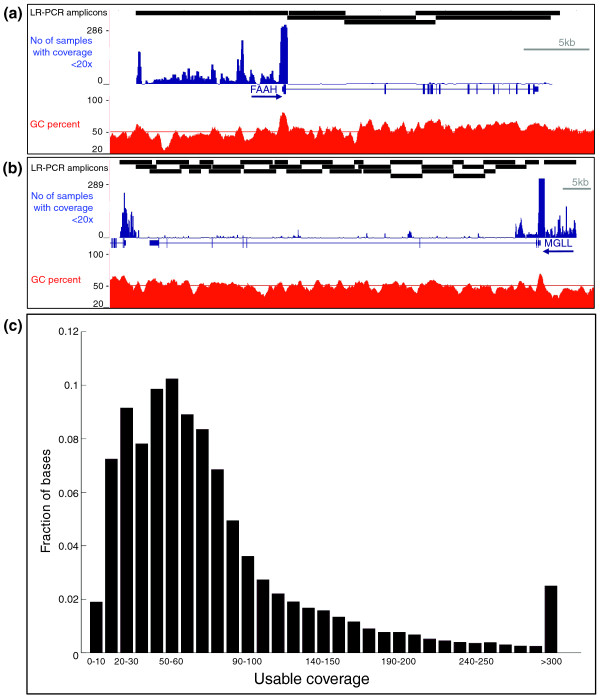
Figure 2.
Sequence coverage distribution. (a,b) Genome Browser tracks showing locations of the 40 LR-PCR amplicons (black rectangles), the number of samples with coverage below 20× (blue histogram, 100-bp windows) and GC percent (red histogram, 10-bp windows) along the FAAH (a) and MGLL (b) re-sequenced intervals. The ends of the intervals have lower coverage due to the fact they were amplified by a single amplicon. The 5' end of the FAAH gene was successfully amplified but coverage is low due to difficulty sequencing high GC content regions. The high GC content at the 5' end of the MGLL gene resulted in an inability to successfully design PCR primer pairs despite several attempts. (c) Distribution of the fraction of bases (y-axis) sequenced at increasing usable coverage (x-axis) for sequence-based association studies. Usable coverage is defined at each base as the minimum coverage reached by 90% or more of the samples.