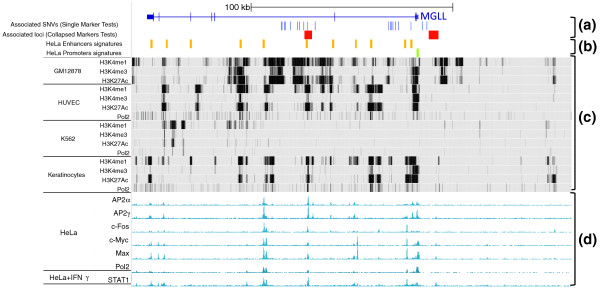
Figure 5.
Functional annotation of the associated variants in the MGLL interval. Track A: variants identified by the single marker tests (blue bars) or the merge of the locus-variants identified by the collapsed marker test RareCover (red boxes) are indicated. Track B: predicted intervals for promoters (green) or transcriptional enhancers (orange) in HeLa cells [45]. Tracks C and D: the distribution of chromatin binding proteins from the ENCODE data obtained from the UCSC genome browser (15 November 2009). Track C: Broad/MGH ENCODE group chromatin signatures corresponding to enhancers (H3K4me1, H3K27ac) and promoters (H3K4me1+3, Pol2) in various cell types (GM12878, HUVEC, K562, Keratinocytes) [46]. Track D: Yale/UCD/Harvard ENCODE group identified AP2α and γ, c-Myc, Max, c-Fos and Pol2 binding sites in HeLa cells, and STAT1 binding sites in HeLa treated with IFNγ [46].