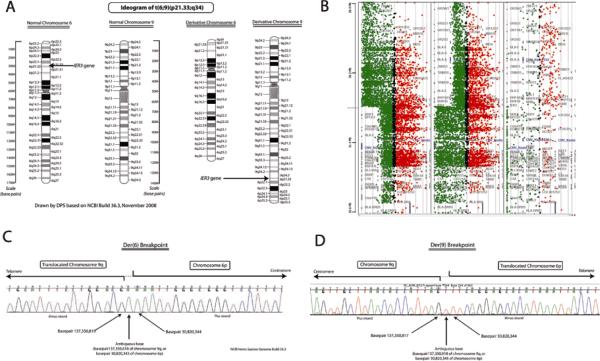
Figure 1.
A, Ideogram of t(6;9)(p23.1;q34), shown to rearrange IER3, in a patient with MDS. Normal chromosomes 6 and 9 are depicted on the left; derivative chromosomes seen in the propositus are depicted on the right. B, Array comparative genomic hybridization (CGH) demonstrating change in DNA copy number for chromosome 6p, centered on IER3, in a patient with MDS. Sample DNA is labeled green, representing intensity of fluorescein signal (proportional to DNA copy number), control DNA is labeled red, representing intensity of Texas Red signal. The IER3 gene is the closest to the apparent breakpoint on chromosome 6p. Array CGH narrowed the breakpoint to within an 800 bp region on chromosome 6p (which included the IER3 transcript initiation site) and 13 kbp on chromosome 9q. C and D, Sequencing chromatogram of breakpoints on derivative chromosome 6p (C) and derivative chromosome 9q (D). Sequencing-defined breakpoints were at base pairs 137,350,819 on chromosome 9 and 30,820,344 on chromosome 6 (NCBI build 36.3), with 1 base pair ambiguity due to a repeated nucleotide. Sequencing was performed in both directions. Southern blotting confirmed that this was a simple, balanced reciprocal translocation.