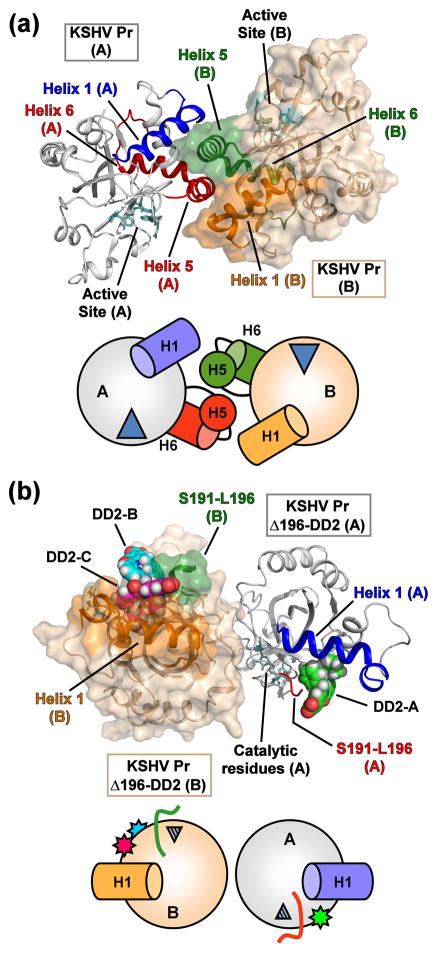
Fig. 3. Structural comparison of the “apo” and DD2-inhibited KSHV Pr monomers.
(a) The dimer structure of peptide-phosphonate inhibited KSHV Pr (2PBK). The catalytic residues (cyan) are located ~ 15 – 20 Å from the dimer interface. The interfacial helix 5 and the following helix 6 (monomer A, red; monomer B, green) are displayed. Helix 1 of monomer A (blue) and monomer B (orange) also form a portion of the dimer interface and are aligned in an anti-parallel orientation with respect to each other. (b) The structure of the KSHV Pr Δ196-DD2 complex (3NJQ) crystallizes as an asymmetric dimer, with dimerization occurring on the opposite face with respect to 2PBK. DD2 molecules bind to the hydrophobic surface normally occupied by helix 5. Monomer A of the complex contains one DD2 molecule (pose 1, green carbons), while monomer B contains two DD2 molecules (pose 2, magenta carbons; pose 3, cyan carbons). The truncated C-terminal residues of the Δ196 constructs (red, monomer A; green, monomer B) are also indicated. Helix 1 of monomer A (blue) and monomer B (orange) are oriented end-on-end with respect to each other. Below each structure are cartoon representations of the monomeric units, with the wedges representing the active site, and stars the DD2 molecules.