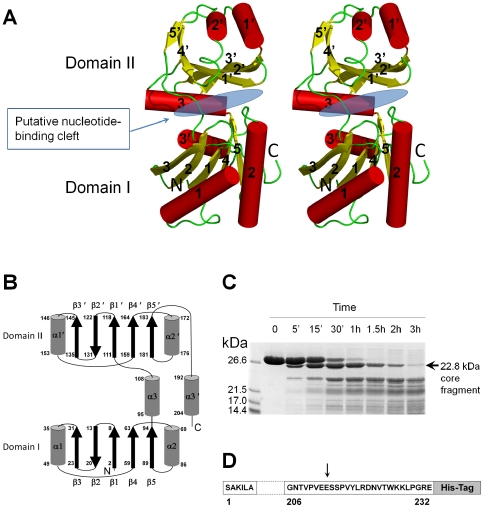
Figure 1. A: Stereo diagram of the structure of the VpYeaZ monomer.
Each element of the secondary structure is labeled. Domains I and II and the putative nucleotide-binding cleft are identified. The figure was prepared using PyMOL [11]. B: The topology of the secondary structure elements. Residue numbers are indicated at the start and end of each secondary structure element. C: SDS-PAGE showing the time-course of VpYeaZ digestion by Glu-C protease. The positions of molecular mass markers are shown to the left. The arrow indicates a relatively stable C-terminally truncated fragment. D: Amino acid sequence of VpYeaZ with Glu-C-sensitive site identified in this study shown by the arrow.