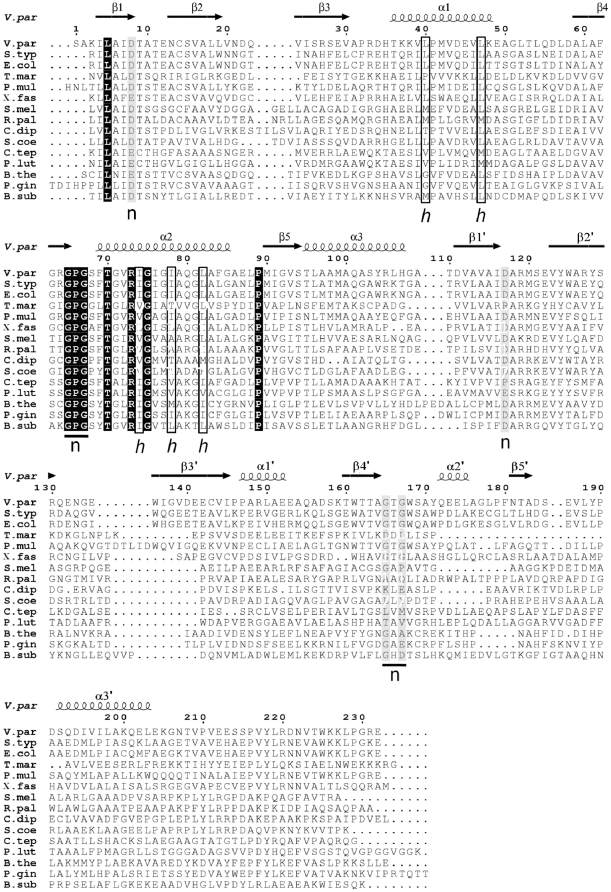
Figure 3. A sequence alignment of YeaZ homologues in V. parahaemolyticus (V. par), S. typhimurium (S. typ), E. coli (E. col), T. maritima (T. mar), Pasteurella multocida (P. mul), Xylella fastidiosa (X. fas), Sinorhizobium meliloti (S. mel), Rhodopseudomonas palustris (R. pal), Corynebacterium diphtheria (C. dip), Streptomyces coelicolor (S. coe), Chlorobium tepidum (C. tep), Pelodictyon luteolum (P. lut), Bacteroides thetaiotaomicron (B. the), Porphyromonas gingivalis (P. gin) and Bacillus subtilis (B. sub).
The elements of the secondary structure and the sequence numbering for VpYeaZ are shown above the alignment. The fully conserved residues are shown by a black box with reverse type. The position of the conserved and semi-conserved residues which can be identified in the nucleotide-binding site of proteins belonging to the ASKHA family, are highlighted by black and gray boxes respectively, and marked with “n” underneath the aligned sequences. The residues forming a hydrophobic surface patch which becomes exposed upon dimer re-organisation are enclosed by a box and marked with “h” underneath the alignment. Alignment was carried out using the ClustalW server [16]. The figure was created using ESPript [17].