Figure 1.
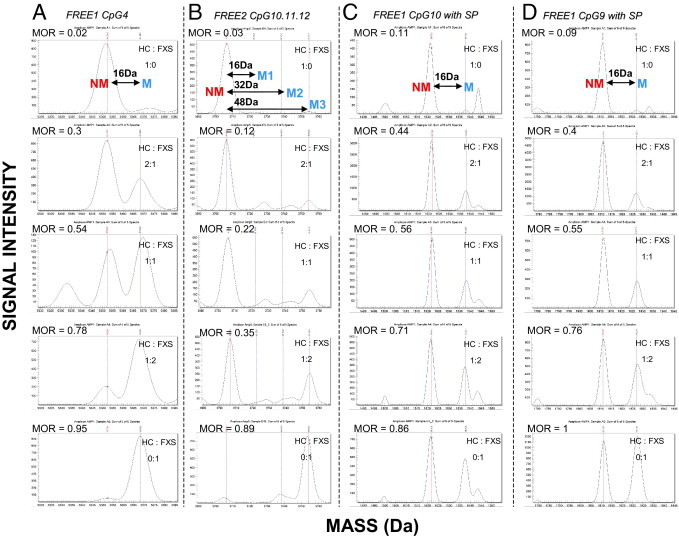
Examples of raw mass spectra for selected FREE1 and FREE2 CpG sites from spiked samples. The mass difference between the NM and M fragments for the same CpG site is 16 Da because of the presence of an adenosine residue in the place of a guanosine residue. The EpiTYPER-used software identifies peaks acceptable for analysis, which is presented in the form of the MOR.29–31 This ratio represents the relationship between M and NM peaks for the same fragments, which is analogous to total percentage of methylation.26 Herein, we present raw spectra for spiked samples included during each run used to indicate if there were any technical problems between different plates/runs and whether the results were comparable between the runs. Lymphoblast DNA from an HC male was spiked with lymphoblast DNA form a male affected with FXS at 1:0, 2:1, 1:1, 1:2, and 0:1 ratios, corresponding to 0%, 33.3%, 50%, 66.6%, and 100% (top to bottom), respectively, FXS DNA in the sample. The peaks representing fragments from UM CpG units are represented by red lines, whereas the fragments from methylated CpG units are represented by blue lines. A: CpG4 fragments with no silent peaks. B: FREE2 CpG 10, 11, and 12 fragments of the same size, with the third blue peak representing the combined mass of methylation fragments from all three sites (54 Da away from the UM peak). C: FREE1 CpG 10 fragments with a silent peak contribution evident (bottom panel). D: FREE1 CpG 9 fragments with a silent peak contribution evident (bottom panel). The corresponding MORs from these raw spectra are plotted against FXS DNA input (see Supplemental Figures S2 and S3 at http://jmd.amjpathol.org). MOR indicates methylation output ratio.
