Abstract
The analysis of X chromosome inactivation (XCI) patterns is a widely used diagnostic tool in clinical practice when investigating X-linked diseases. The most commonly used assay to determine XCI patterns takes advantage of a locus within the androgen receptor (AR) gene. This PCR-based assay relies on two differentially methylated restriction enzyme sites (HpaII) and a polymorphic repeat located within this locus. Although highly informative, this locus is not always sufficient to evaluate the X-inactivation status in X-linked disorders. We have identified three new loci that can be used to determine XCI patterns in a methylation-sensitive PCR-based assay. All three loci contain polymorphic repeats and a methylation-sensitive restriction enzyme (HpaII) site, methylation of which was shown to correlate with XCI. DNA from 60 females was used to estimate the heterozygosity of these new loci. The reliability of the loci was validated by showing a high correlation between the results obtained by employing the new loci and the AR locus using DNA from 15 females who were informative for all four loci. Altogether, we show that these loci can be applied easily in molecular diagnostic laboratories, either as a supplement or as an alternative to the existing AR assay.
X chromosome inactivation (XCI) in females is a mechanism of dosage compensation that equalizes the expression of X-linked genes between the sexes.1 Normally, XCI is a random process in which the maternal and the paternal X chromosome have the same chance of being inactivated in each cell. As a consequence, females are mosaic for two cell populations differing in the parental origin of the expressed X chromosome. In most females, these two cell populations are represented equally; however, skewing of XCI patterns is observed in about 10% of the female population (reviewed by Orstavik2). Importantly, skewing is known to influence the clinical manifestation of a number of X-linked diseases.3,4 Therefore, it is essential to investigate XCI patterns in female members of families with X-linked disorders to determine carrier status and provide optimal genetic counseling.2
Several assays used for determining XCI patterns have been published. These are normally based on Southern blot hybridization or PCR. Assays relying on Southern blot hybridization (eg, HPRT5) are labor-intensive, and assays based on restriction fragment length polymorphism (eg, PGK16) are less informative as compared with short, tandem repeat PCR-based assays. Some of these PCR-based assays, however, may pose problems because of other reasons such as difficulties in PCR amplification caused by a GC-rich repeat (eg, FMR17), requirement of several enzyme digestion steps (eg, MAOA8), or complicated quantification owing to shadow peaks (eg, ZNF2619).
The most commonly used short, tandem repeat–based assay takes advantage of two methylation-sensitive restriction enzyme sites (HpaII) and a polymorphic CAG repeat located within exon 1 of the androgen receptor (AR) gene.10 Both restriction sites are methylated on the inactive X chromosome. Up to 90% of the female population is heterozygous for the CAG repeat,11,12 but in clinical practice uninformative families are encountered occasionally and this necessitates availability of alternative methods.
In this study, we describe three new methylation-sensitive PCR-based assays using three different loci along the X chromosome.
Materials and Methods
In Silico Search for New Loci
The search for new loci was performed using the University of Santa Cruz genome browser (http://genome.ucsc.edu) and relies on the fact that CpG islands in the promoter regions of X-linked genes generally are hypermethylated on the inactive X chromosome compared with those on the active X chromosome.13 We screened the whole X chromosome for loci that concurrently included a methylation-sensitive restriction enzyme site located within or close to a promoter CpG island of an X-linked gene, and a polymorphic repeat. Three loci that fulfilled these requirements were selected for further investigations.
Subjects and DNA Isolation
DNA was isolated using standard methods from five males and 60 Caucasian females not known to have an X-linked disease, two female carriers of Xq28 duplication, and one female carrying a partial deletion of the NEMO gene (NF-κB essential modulator). The procedures involving DNA samples were conducted according to the Declaration of Helsinki.
HpaII Digestion of DNA
DNA (2 μg) was digested with 20 U HpaII (New England Biolabs, Ipswich, MA) in a total reaction volume of 20 μL and incubated at 37°C for at least 12 hours.
PCR Amplification and Fragment Analysis
PCR amplification was performed using 100 ng (from females) or 200 ng (from males) undigested DNA or 2 μL of HpaII digested DNA. The primers and PCR conditions are listed in Table 1 and the PCR reagents are listed in Table 2. The PCR fragment sizes for the most frequent allele of the three loci were as follows: 539 bp for the zinc finger DHHC-type containing 15 (ZDHHC15) locus (21 repeats), 392 bp for SLIT and NTRK-like family, member 4 (SLITRK4) locus (25 repeats), and 334 bp for proprotein convertase subtilisin/kexin type 1 inhibitor (PCSK1N) locus (31 repeats).
Table 1.
PCR Primers and Conditions Used to Amplify the ZDHHC15, SLITRK4, PCSK1N, and AR Loci
| Locus | Primers | PCR conditions |
|---|---|---|
| ZDHHC15 | Forward⁎: 5′-TCTTTGGCTCGAAGATCGAC-3′ | 95°C/45 seconds |
| Reverse: 5′-TATGGCTCGCATCTTTCACA-3′ | (95°C/45 seconds; 58°C/45 seconds; 72°C/45 seconds) 23×72°C/10 minutes | |
| SLITRK4 | Forward⁎: 5′-GCACACAAGCAGTCCTTCCT-3′ | 95°C/15 minutes |
| Reverse: 5′-TGGCTTCTTGGTTGCTCTCT-3′ | (94°C/45 seconds; 55°C/45 seconds; 72°C/1 minute) 23×72°C/10 minutes | |
| PCSK1N | Forward⁎: 5′-ATGCGAAGACCATTCCCTCT-3′ | 95°C /10 minutes |
| Reverse: 5′-GTGCGTGTGATGTGAGGAGA-3′ | (95°C/45 seconds; 58°C/45 seconds; 72°C/45 seconds) 22×72°C/7 minutes | |
| AR | Forward⁎: 5′-TCCAGAATCTGTTCCAGAGCGTGC-3′ | 95°C/10 minutes |
| Reverse: 5′-GCTGTGAAGGTTGCTGTTCCTCAT-3′ | (95°C/45 seconds; 58°C/45 seconds; 72°C/45 seconds) 25×72°C/7 minutes |
Primers marked with a FAM tag.
Table 2.
PCR Reagents Used to Amplify the ZDHHC15, SLITRK4, PCSK1N, and AR Loci
| Locus | Total volume (μL) | Polymerase | Buffer | dNTPs (μmol/L) | Primer (μmol/L) |
|---|---|---|---|---|---|
| ZDHHC15 | 50 | 2.5 U Cloned Pfu⁎ | Cloned Pfu buffer × 1⁎ | 200 | 0.5 |
| SLITRK4 | 20 | 0.5 U HotStarTaq polymerase† | Buffer × 1† | 200 | 0.5 |
| PCSK1N | 20 | 1 U AmpliTaq Gold polymerase‡ | Buffer × 1 included MgCl2‡ | 200 | 0.5 |
| AR | 20 | 1 U AmpliTaq Gold polymerase‡ | Buffer × 1 included MgCl2‡ | 200 | 0.5 |
Buffers, nucleotides, primers, and MgSO4 are given in final concentrations.
Stratagene (Santa Clara, CA),
Qiagen (Germantown, MD),
Applied Biosystems (Foster City, CA).
Forward primers were labeled with a FAM-tag and PCR products were analyzed on an ABI 3130 XL genetic analyzer (Applied Biosystems, Foster City, CA) using GeneScan 500 ROX (Applied Biosystems) as an internal-lane size standard and GeneMapper software (Applied Biosystems). The X-inactivation ratios were calculated as previously described,14 X-inactivation was defined as being skewed if more than 80% of the investigated cells inactivated the same X chromosome, and extremely skewed if the ratio was higher than 90%.
Heterozygosity and Polymorphism Information Content Value
The expected heterozygosity (HT) of each locus was calculated with the following:
and the polymorphism information content value (PIC) was calculated with the following:
where pi and pj are the observed frequencies of the ith and jth alleles at a given locus. The results were obtained by use of the calculation function available at http://www.chrx-str.org.
Results and Discussion
In this study we describe three new loci that can be used in determining X-inactivation patterns. All loci contain one or two polymorphic dinucleotide repeats and a restriction site for the methylation-sensitive enzyme HpaII (see Supplemental Figure S1 at http://jmd.amjpathol.org). The first locus is localized immediately upstream to the first exon of the ZDHHC15 gene at Xq13.3 and contains an AC dinucleotide repeat. The second locus is within the promoter region of the SLITRK4 gene at Xq27.3 and it includes an AC dinucleotide repeat. The third locus is located at Xp11.23 within the first intron of the PCSK1N gene and harbors both a CA repeat and an AG repeat in tandem.
Correlation between Methylation and X Chromosome Inactivation
The three loci could not be PCR-amplified when using HpaII-digested DNA from 5 male controls (Figure 1A), indicating that the enzyme sites were unmethylated on the active X chromosome. We also investigated two females with Xq28 duplication (Figure 1B, F1 and F3), and one female with partial deletion of the NEMO gene at Xq28 (Figure 1B, F2) using these three new loci. All these females were phenotypically normal, suggesting that the X chromosome harboring the aberration was preferentially inactivated, supported by the AR assay, which showed extremely skewed XCI. The female patients (F1, F2, and F3) were heterozygous for the ZDHHC15, SLITRK4, and PCSK1N loci, respectively. By using HpaII-digested DNA from these females, only a single allele of their respective heterozygous loci could be amplified with PCR (Figure 1B). These results suggest that the HpaII sites within these loci are methylated on the inactive X chromosome, similar to the AR locus.
Figure 1.
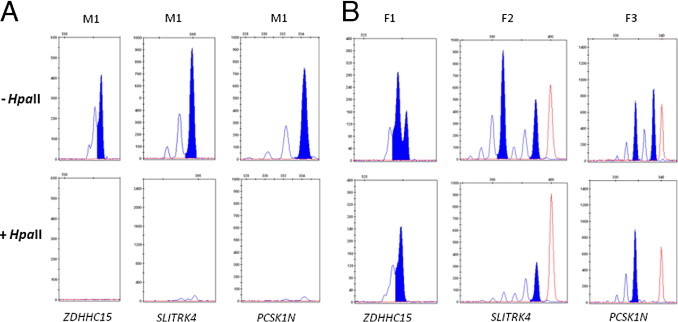
Fragment analysis results for the ZDHHC15, SLITRK4, and PCSK1N loci. Undigested (−HpaII) or digested (+HpaII) DNA was amplified with PCR, using the respective primer sets for the three loci. The fragment analysis was performed with GeneMapper software (Applied Biosystems). A: After HpaII digestion, DNA from the male individual (M1) cannot be amplified with PCR. B: Three phenotypically normal females (F1–F3) with extremely skewed X chromosome inactivation. F1 and F3 have Xq28 duplication and F2 has partial deletion of the NEMO gene. As seen on the lower panel, one of the alleles (as represented by PCR fragments, blue peaks) cannot be amplified by PCR after HpaII digestion. The red peaks represent the size standard 500 ROX.
Heterozygosity and Polymorphism Information Content Values of the New Loci
The heterozygosity of the loci was investigated in 60 Caucasian females. Of these, 35 (58%) were heterozygous for the ZDHHC15 locus, 48 (80%) were heterozygous for the SLITRK4 locus, and 45 (75%) were heterozygous for the PCSK1N locus. We determined the number of repeat units for different alleles by sequencing the most frequent alleles and estimated the allele frequencies. The expected heterozygosity was calculated and the polymorphism information content values of the ZDHHC15, PCSK1N, and SLITRK4 loci were determined to be 0.55, 0.64, and 0.82, respectively. Information on the loci, including number of repeat units, allele frequency, observed and expected heterozygosity values, and polymorphism information content value, are listed in Supplemental Table S1 (available at http://jmd.amjpathol.org).
Comparison between Results Obtained by Use of the AR Locus and the Three New Loci
We compared the methylation status of the three new loci with the AR locus in 15 females heterozygous for all these loci (Table 3). DNA from a male was used as an indirect control for complete HpaII digestion. In 8 of 15 females (no. 1, 4, 5, 7, 10, 12, 14, and 15) there was a clear correlation between the XCI ratios for all of the loci. In five females (no. 2, 6, 9, 11, and 13), the XCI ratios were correlated in three of the four loci. Generally, none of the loci was more discrepant than the others and the discrepancies observed may be owing to different reasons as explained later.
Table 3.
XCI Ratios of 15 Females Using the AR, ZDHHC15, SLITRK4, and PCSK1N Loci
| Female no. | AR | ZDHHC15 | SLITRK4 | PCSK1N |
|---|---|---|---|---|
| 1 | 25:75 | 22:78⁎ | 20:80 | 18:82 |
| 2 | 27:73⁎ | 29:71 | 15:85 | 24:76 |
| 3 | 28:72 | 49:51⁎ | 21:79 | 42:58⁎ |
| 4 | 48:52 | 45:55 | 47:53⁎ | 47:53⁎ |
| 5 | 49:51 | 49:51 | 40:60 | 49:51 |
| 6 | 33:67 | 13:87 | 30:70 | 32:68 |
| 7 | 45:55 | 44:56⁎ | 50:50 | 48:52 |
| 8 | 24:76 | 29:71⁎ | 44:56 | 44:56⁎ |
| 9 | 46:54 | 49:51 | 42:58 | 30:70 |
| 10 | 42:58⁎ | 47:53⁎ | 45:55 | 39:61 |
| 11 | 37:63 | 47:53 | 44:56 | 46:54 |
| 12 | 44:56 | 46:54 | 49:51 | 46:54 |
| 13 | 46:54⁎ | 36:64⁎ | 44:56 | 46:54 |
| 14 | 49:51⁎ | 50:50⁎ | 49:51 | 47:53 |
| 15 | 47:53⁎ | 46:54 | 46:54 | 46:54 |
Loci where the two alleles are separated by only a single repeat unit.
The presence of shadow peaks occasionally creates difficulties in interpreting allele ratios in PCR-based methodologies. Shadow peak artifacts arise during PCR amplification and give rise to products containing one repeat unit less than the template DNA. Thus, when the two alleles of a locus are separated only by a single repeat unit (Table 3) the shadow peak of the longer allele will superimpose the peak of the shorter allele and influence the interpretation of the result. Some of the discrepancies observed between the different loci in our study may be explained by the presence of shadow peaks. The use of several different loci simultaneously in determining X-inactivation patterns will therefore be an advantage in overcoming the general problem with shadow peaks.
Methylation-based methods depend on the methylation state of a single or a few cytosine residues. Thus, if the methylation state of the cytosine does not completely correlate with the activity status of the X chromosome in a single individual, the method will give rise to an incorrect XCI ratio. Discrepancies between XCI ratios obtained by different assays, including the AR and FMR1 assays, previously have been described in several studies.9,15 Thus, in some individuals the methylation of a single cytosine residue might not be representative for the inactivation status of the whole X chromosome. This could explain the observed discrepancies between the different loci in our study. The use of several loci simultaneously therefore may be a more reliable approach.
In this study we introduce three loci, ZDHHC15, SLITRK4, and PCSK1N, which may be used in XCI studies as a supplement or an alternative to the AR assay. We also suggest using several loci to determine the X chromosome inactivation status of heterozygote females to eliminate interindividual methylation differences.
Footnotes
K.R. and Z.T. contributed equally to this work.
Supplemental material for this article can be found at http://jmd.amjpathol.org or at doi: 10.1016/j.jmoldx.2011.05.003.
Supplementary data
Location of the three new loci. The location of each locus is indicated on the X chromosome. A: The ZDHHC15 locus is located just upstream of the first exon of the ZDHHC15 gene and contains an AC dinucleotide repeat. B: The SLITRK4 locus is located upstream of the first exon of the SLITRK4 gene and contains an AC dinucleotide repeat. C: The PCSK1N locus is located upstream of the first exon of the PCSK1N gene and contains a CA repeat and an AG repeat in tandem. Each locus includes a single restriction site of the methylation-sensitive enzyme HpaII indicated by a red cross. The figure is adapted from the UCSC Genome Browser (NCBI36/hg18 assembly).
References
- 1.Lyon M.F. X-chromosome inactivation as a system of gene dosage compensation to regulate gene expression. Prog Nucleic Acid Res Mol Biol. 1989;36:119–130. doi: 10.1016/s0079-6603(08)60166-x. [DOI] [PubMed] [Google Scholar]
- 2.Orstavik K.H. X chromosome inactivation in clinical practice. Hum Genet. 2009;126:363–373. doi: 10.1007/s00439-009-0670-5. [DOI] [PubMed] [Google Scholar]
- 3.Nelson D.L. NEMO, NFkappaB signaling and incontinentia pigmenti. Curr Opin Genet Dev. 2006;16:282–288. doi: 10.1016/j.gde.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 4.Redonnet-Vernhet I., Ploos van Amstel J.K., Jansen R.P., Wevers R.A., Salvayre R., Levade T. Uneven X inactivation in a female monozygotic twin pair with Fabry disease and discordant expression of a novel mutation in the alpha-galactosidase A gene. J Med Genet. 1996;33:682–688. doi: 10.1136/jmg.33.8.682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tefferi A., Thibodeau S.N., Solberg L.A., Jr Clonal studies in the myelodysplastic syndrome using X-linked restriction fragment length polymorphisms. Blood. 1990;75:1770–1773. [PubMed] [Google Scholar]
- 6.Gilliland D.G., Blanchard K.L., Levy J., Perrin S., Bunn H.F. Clonality in myeloproliferative disorders: analysis by means of the polymerase chain reaction. Proc Natl Acad Sci U S A. 1991;88:6848–6852. doi: 10.1073/pnas.88.15.6848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee S.T., McGlennen R.C., Litz C.E. Clonal determination by the fragile X (FMR1) and phosphoglycerate kinase (PGK) genes in hematological malignancies. Cancer Res. 1994;54:5212–5216. [PubMed] [Google Scholar]
- 8.Hendriks R.W., Chen Z.Y., Hinds H., Schuurman R.K., Craig I.W. An X chromosome inactivation assay based on differential methylation of a CpG island coupled to a VNTR polymorphism at the 5′ end of the monoamine oxidase A gene. Hum Mol Genet. 1992;1:187–194. doi: 10.1093/hmg/1.3.187. [DOI] [PubMed] [Google Scholar]
- 9.Beever C., Lai B.P., Baldry S.E., Peñaherrera M.S., Jiang R., Robinson W.P., Brown C.J. Methylation of ZNF261 as an assay for determining X chromosome inactivation patterns. Am J Med Genet A. 2003;120A:439–441. doi: 10.1002/ajmg.a.20045. [DOI] [PubMed] [Google Scholar]
- 10.Allen R.C., Zoghbi H.Y., Moseley A.B., Rosenblatt H.M., Belmont J.W. Methylation of HpaII and HhaI sites near the polymorphic CAG repeat in the human androgen-receptor gene correlates with X chromosome inactivation. Am J Hum Genet. 1992;51:1229–1239. [PMC free article] [PubMed] [Google Scholar]
- 11.Edwards A., Hammond H.A., Jin L., Caskey C.T., Chakraborty R. Genetic variation at five trimeric and tetrameric tandem repeat loci in four human population groups. Genomics. 1992;12:241–253. doi: 10.1016/0888-7543(92)90371-x. [DOI] [PubMed] [Google Scholar]
- 12.Sleddens H.F., Oostra B.A., Brinkmann A.O., Trapman J. Trinucleotide repeat polymorphism in the androgen receptor gene (AR) Nucleic Acids Res. 1992;20:1427. doi: 10.1093/nar/20.6.1427-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salstrom J.L. X-inactivation and the dynamic maintenance of gene silencing. Mol Genet Metab. 2007;92:56–62. doi: 10.1016/j.ymgme.2007.05.015. [DOI] [PubMed] [Google Scholar]
- 14.Lau A.W., Brown C.J., Peñaherrera M., Langlois S., Kalousek D.K., Robinson W.P. Skewed X-chromosome inactivation is common in fetuses or newborns associated with confined placental mosaicism. Am J Hum Genet. 1997;61:1353–1361. doi: 10.1086/301651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Busque L., Mio R., Mattiolo J., Brais E., Blais N., Lalonde Y., Maragh M., Gilliland D.G. Nonrandom X-inactivation patterns in normal females: lyonization ratios vary with age. Blood. 1996;88:59–65. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Location of the three new loci. The location of each locus is indicated on the X chromosome. A: The ZDHHC15 locus is located just upstream of the first exon of the ZDHHC15 gene and contains an AC dinucleotide repeat. B: The SLITRK4 locus is located upstream of the first exon of the SLITRK4 gene and contains an AC dinucleotide repeat. C: The PCSK1N locus is located upstream of the first exon of the PCSK1N gene and contains a CA repeat and an AG repeat in tandem. Each locus includes a single restriction site of the methylation-sensitive enzyme HpaII indicated by a red cross. The figure is adapted from the UCSC Genome Browser (NCBI36/hg18 assembly).


