Abstract
The SPO11 protein generates programmed DNA double-strand breaks (DSBs) that initiate meiotic recombination. Endonucleolytic cleavage 3′ to the DSB sites releases SPO11 from DNA, leaving SPO11 covalently associated with an oligonucleotide. This chapter describes detection of the release product, SPO11-oligonucleotide complexes, from mouse testis lysates. The method for determining the size of SPO11-associated oligonucleotides is also provided.
Keywords: SPO11, mouse, testes, terminal transferase, DNA double-strand break
1. Introduction
To detect mouse SPO11-oligonucleotide complexes, SPO11 is immunoprecipitated from testis lysates and immunoprecipitates are labeled with terminal transferase (TdT) in the presence of radioactive nucleotides. This protocol is largely derived from the corresponding protocol from budding yeast (Chapter 12 from this volume). Unlike synchronous meiotic cultures of yeast, only a small fraction of cells in adult testes are spermatocytes in early meiotic prophase (1), the stage in which SPO11-oligonucleotide complexes are likely to be present based on results from budding yeast (2). In addition, the amount of SPO11-oligonucleotide complexes is likely to be comparable per meiotic cell in mouse and budding yeast due to similar numbers of SPO11-induced double-strand breaks (3), but the mouse genome (~2.5 × 109 bp) is about 200 times the size of the budding yeast genome (~1.2× 107 bp). The small amount of SPO11-oligonucleotide complexes versus the large amount of genomic DNA in mouse poses a challenge for detection. To increase the signal to noise ratio, a simple step of ultracentrifugation is used to pellet most genomic DNA from crude testis lysates, and SPO11-associated oligonucleotide complexes are then immunoprecipitated from the supernatant. To determine the size of SPO11-associated oligonucleotides, SPO11-oligonucleotide complexes are purified using SDS-PAGE and then deproteinized. DNA is then precipitated and resolved on a sequencing gel.
2. Materials
2.1. Preparation of testis lysates
4 1/2” and 3 1/2” dissecting scissors, 2 fine-pointed dissection forceps.
PBS: 137 mM NaCl, 2.7 mM KCl, 10 mM NaH2PO4 / Na2HPO4, pH 7.4.
Lysis buffer: 1% Triton X-100, 400 mM NaCl, 25 mM HEPES-NaOH, pH 7.4, 5 mM EDTA. Supplement with Complete, Mini, EDTA-free protease inhibitor cocktail (Roche; use 1 tablet per 10 mL lysis buffer) and 2 mM dithiothreitol immediately before use.
Plastic pestles to fit 1.5 ml centrifuge tubes.
Benchtop ultracentrifuge (or floor ultracentrifuge for large scale).
Beckman TLA100.2 rotor or equivalent for 1 mL-capacity ultracentrifuge tubes (or for large scale, Sorvall AH-650 rotor or equivalent for 3.5 mL or 5 mL ultracentrifuge tubes).
1 mL-capacity polycarbonate ultracentrifuge tubes (or for large scale, 3.5-mL or 5.5-mL capacity ultracentrifuge tubes).
2.2. Immunoprecipitation
Antibody against mouse SPO11 (Kamiya Biomedical Company, clone 129/180).
Protein A agarose beads (Roche).
Wash buffer: 1% Triton X-100, 150 mM NaCl, 15 mM Tris-HCl, pH 7.4.
2.3. TdT Labeling
Terminal transferase (FPLCpure from GE healthcare, or recombinant from New England Biolabs).
10× Labeling buffer (equivalent to 10× NEB4 from New England Biolabs): 20 mM Tris-acetate, pH 7.9, 50 mM potassium acetate, 10 mM magnesium acetate, 1 mM dithiothreitol.
[α-32P] dCTP (6000 Ci/mmol, 20 mCi/mL).
2.4. Detection of labeled mouse SPO11-oligonucleotide complexes
2× SDS-PAGE sample buffer: 100 mM Tris-HCl, pH 6.8, 4% SDS, 20% glycerol, 10% β-mercaptoethanol, 10% saturated bromophenol blue.
SDS-PAGE reagents and apparatus.
PVDF or nitrocellulose membrane.
CAPS buffer: 10 mM cyclohexylaminopropane sulfonic acid, pH 11, 10% methanol. Adjust 100 mM CAPS stock to pH 11 with NaOH. To make CAPS buffer, mix 1 vol of 100 mM CAPS stock, 1 vol of methanol, and 8 vol of water.
Phophor imaging plate and analyzer.
2.5. Detection of SPO11 protein by Western blotting
PBST: PBS containing 0.1% Tween 20.
Blocking solution: 2% BSA and 0.05% sodium azide in PBST.
Horse radish peroxidase (HRP) -conjugated anti-mouse secondary antibody (GE healthcare or equivalent).
ECL plus Western blotting detection system (GE healthcare).
2.6. Determination of the length of SPO11-associated oligonucleotides
Low-retention centrifuge tubes, 2 mL and 1.5 mL.
Protease K elution mix: 50 mM Tris-HCl, pH 8.0, 0.5% SDS, 1 mM EDTA, 1 mM CaCl2, 100 µg/mL protease K.
9 M ammonium acetate.
Glycogen.
Absolute ethanol and 70% ethanol (kept at −20°C).
Sequencing gel loading buffer: 80% deionized formamide, 10 mM EDTA, pH 8.0, 0.5 mg/mL xylene cyanol FF, 10% saturated bromophenol blue.
Sequencing gel reagents and apparatus.
10 bp DNA ladder (Invitrogen).
T4 polynucleotide kinase.
[γ-32P] ATP (3000 Ci/mmol, 10 mCi/mL).
0.4 M dithiothreitol.
Sequencing gel fixing solution: 10% methanol, 7% acetic acid in water.
Cellulose paper, DE81 paper.
3. Methods
3.1. Preparation of testis lysates
Prepare four wild-type mice (for positive and negative controls) and two mice from other strains of interest (see Note 1). Sacrifice animals using carbon dioxide and collect testes in ice-cold PBS.
Rinse testes with PBS a few times. Tear apart and remove the outer membrane (tunica albuginea) using two fine-pointed dissection forceps. Drop de-capsulated testes into 1.5 mL tubes placed on ice, one testis per tube.
Add 450 µL cold lysis buffer to each tube. Grind testes with plastic pestles. About half a minute of grinding is needed to obtain a viscous crude lysate without visible cell clumps (see Note 2).
Immediately transfer crude lysates into cold, 1 mL-capacity ultracentrifuge tubes. Each tube holds lysates from two testes. Use P1000 pipet tips with about 3 mm removed from ends to pipet the viscous lysates.
Balance the centrifuge tubes using lysis buffer. Spin the crude lysates at 100,000 rpm (355,040g) in a TLA 100.2 rotor (or equivalent) on a benchtop ultracentrifuge for 15 min at 4°C. Pellets contain insoluble material and most of the genomic DNA. Carefully decant supernatants into 15 mL tubes, one strain per tube (see Note 3).
3.2. Immunoprecipitation
Divide cleared lysate from wild-type animals equally into two 15 mL tubes. To one tube of wild-type lysate, add 40 µL PBS (mock immunoprecipitation as the negative control). To the other tube of wild type lysate (positive control), add 40 µL SPO11 antibody (200 µg/mL, use at 2 µg/ testis). Also add 40 µL SPO11 antibody to lysates from other strains. Incubate all reactions at 4°C with end-over-end rotation for 1 h (see Note 4).
Add 60 µL slurry of protein A agarose beads (15 µL slurry/testis) to all reactions (see Note 5). Incubate at 4°C for 3 h to overnight with end-over-end rotation.
Spin beads down at 3000 rpm (~1700g) for 3 min in a table-top clinical centrifuge. Remove supernatants.
Add 0.5 mL cold wash buffer to each tube. Transfer resuspension into 1.5 mL tubes. Spin beads down in a table-top microcentrifuge at 5000 rpm (~ 2300g) for 1 min. Remove supernatants. Wash beads three times more with cold wash buffer.
3.3. TdT Labeling
Wash beads twice with 300 µL cold 1× labeling buffer. After the last wash, remove as much liquid as possible without disturbing the beads using a P20 pipet.
- Set up TdT reaction mix. Use fresh [α-32P] dCTP. (If the size of SPO11-associated oligonucleotides is to be determined (Subheading 3.6.), use radioactive cordycepin triphosphate for labeling. See Note 6.)
1× reaction mix 10× labeling buffer 5 µL [α-32P] dCTP 2 µL TdT (~20,000 U/mL) 1 µL Water 42 µL Total volume 50 µL Add 50 µL TdT reaction mix to each tube. Incubate at 37°C for 45 min. Mix reactions every 5 min.
Spin beads down. Remove supernatants. Wash beads three times with 0.5 mL cold wash buffer. Remove as much liquid as possible with a P20 pipet after the last wash.
3.4. Detection of labeled mouse SPO11-oligonucleotide complexes
Add 30 µL of 2× SDS sample buffer to each tube. Mix well, incubate at 95°C for 2 min, and chill immediately on ice.
Resolve samples on an 8% SDS-PAGE gel. For each sample, load 20 µL of supernatant excluding beads (about 2.6 testes equivalent). Also load a lane of prestained protein size standard. Stop the run when the bromophenol blue dye front has just migrated out of the gel (about 1 h 8 min at 150 V on a Bio-Rad Mini-protean electrophoresis apparatus).
Transfer the gel to a PVDF or nitrocellulose membrane in CAPS transfer buffer (1 h at 100 V at 4°C on a Bio-Rad Mini-protean electrophoresis apparatus for a 0.75 mm gel) (see Note 7).
Cover the membrane with plastic wrap. Put a piece of Scotch tape over the protein size standard. Make a hot pen by dipping a Sharpie marker into radioactive liquid waste, and use the pen to mark the size standard on the tape. Cover the radioactive marks with another piece of Scotch tape. (If the region containing SPO11-oligonucleotide complexes is to be excised for sizing the oligonucleotides (Subheading 3.6.), also mark the four corners of the membrane with the hot pen for orientation purposes.)
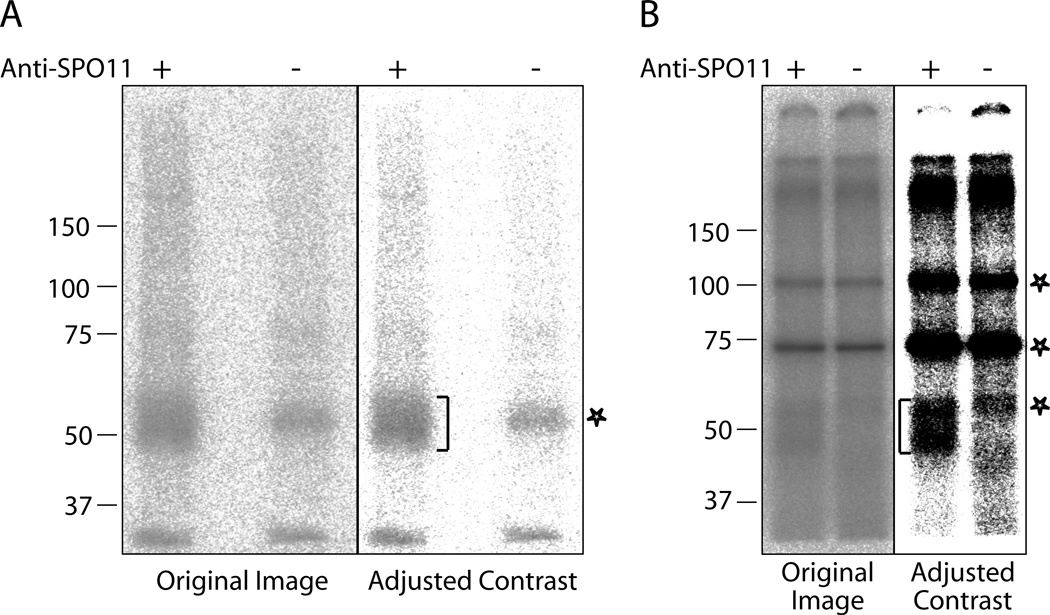
Expose the membrane to a phosphor imaging plate from 4 h to overnight. SPO11-oligonucleotide complexes from wild-type mice migrate between 47–59 kD (Fig. 1) (see Notes 8 and 9).
Fig. 1.
Examples of labeling of mouse SPO11-oligonucleotide complexes. SPO11-oligonucleotide complexes were immunoprecipitated from wild type testis lysate and labeled with TdT and [α-32P] dCTP. Labeling specific to SPO11-oligonucleotide complexes is marked with brackets, and non-specific labeling is marked with asterisks. In both A and B, the left panel is the original image and in the right panel contrast has been adjusted with Photoshop. A. Labeling reactions were resolved on a 7.5% Ready gel (Bio-Rad). Nonspecific labeling at around 55 kD in the lane without SPO11 antibody (mock immunoprecipitation control) is associated with contaminant(s) from the TdT (GE healthcare). Input to lanes: about 2.6 testes equivalent. B. Labeling reactions were resolved on an 8% SDS gel. Nonspecific bands at 75 and 100 kD, as well as the smear at about 60 kD in the lane without SPO11 antibody is associated with contaminants from the TdT (New England Bioloabs). Input to lanes: about 6 testes equivalent.
3.5. Detection of SPO11 protein by Western blotting (skip this section and go directly to Subheading 3.6. if the size of SPO11-oligonucleotide is to be determined.)
Unwrap the membrane and incubate it with blocking solution on a rocking platform for 30 min at room temperature.
Pour off blocking solution and replace with SPO11 antibody diluted 1:500 in blocking solution. Incubate on a rocking platform at room temperature for 1 h.
Wash the membrane 3 times with PBST, 5 min per wash.
Incubate the membrane with HRP-conjugated anti-mouse secondary antibody (for the antibody from GE healthcare, dilute 1:10,000 in PBST containing 5% non-fat dry milk) for 30 min at room temperature.
Wash the membrane 3 times with PBST, 5 min per wash.
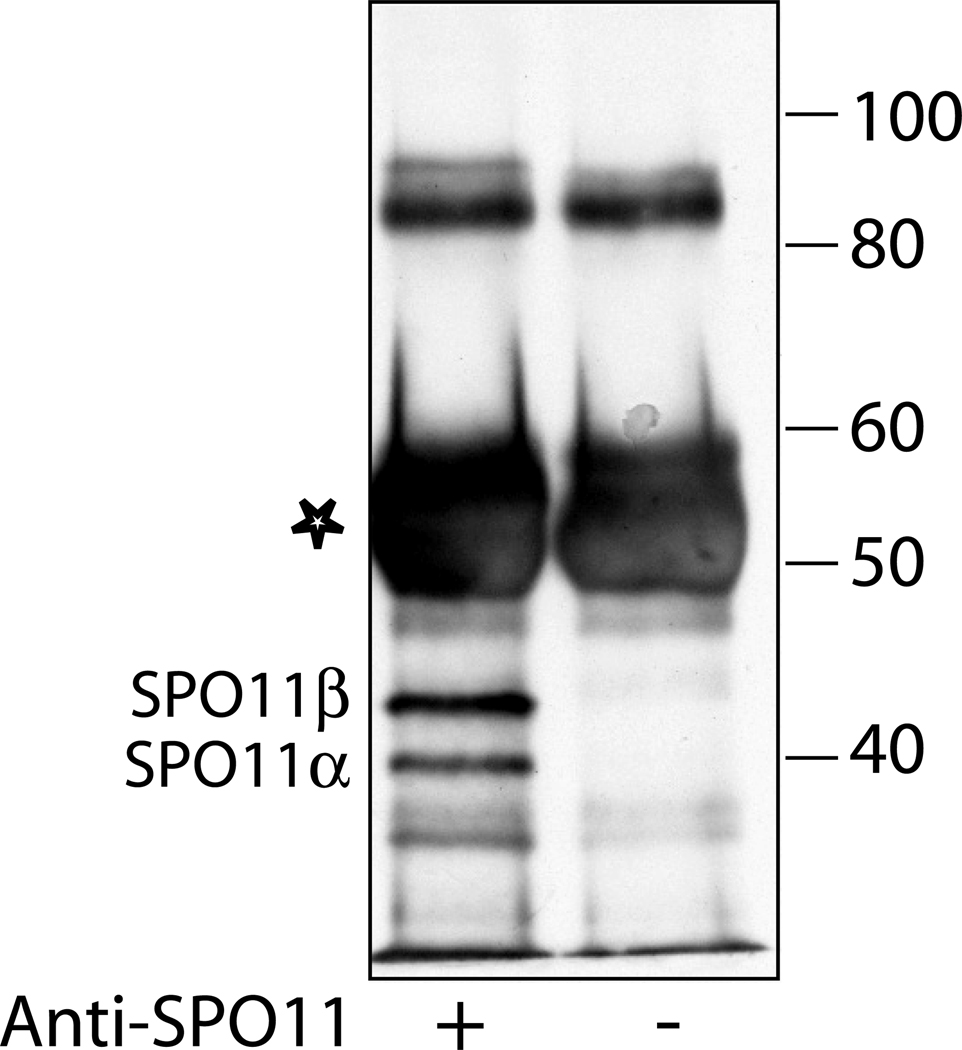
Detect with ECL+ Western blotting detection system according to manufacturer’s instructions. Mouse SPO11 is expressed as multiple isoforms (4, 5). Western blotting detects two bands of free SPO11 migrating at about 40 and 44 kD, respectively (Fig. 2).
Fig. 2.
SPO11 protein detected by Western blotting. Anti-SPO11 or mock immunoprecipitates from wild-type testis lysates were probed with SPO11 antibody. Input to lanes: ~ 2.6 testes equivalent. The antibody heavy chain is marked with an asterisk.
3.6. Determining the length of SPO11-associated oligonucleotides
Print out the image obtained from exposure to the phosphor imaging plate at 100% size. On a light box, align the Sharpie marks on the membrane with the marks on the print-out. Draw a box on the membrane around the regions that contain the SPO11-oligonucleotide complexes, and the corresponding region in the control lane.
Cut out boxed regions, and put these small pieces of membrane into 2 mL low-retention centrifuge tubes, one strain per tube. Add 0.5 mL protease K elution mix to each tube. Seal the tubes with Parafilm. Elute at 50°C with mixing overnight (see Note 10).
Take the membrane out of tubes using clean forceps. Add 150 µl 9 M ammonium acetate, 1 µL 10 mg/mL glycogen, and 1.25 mL absolute ethanol to each tube. Precipitate on dry ice for 1 h. Spin at 13,200g in a microcentrifuge at 4°C for 15 min. Rotate tubes 180°C and spin for another 15 min. Rotate tubes 180°C again and spin for the last 15 min to collect most precipitates at the bottom of the tubes. Remove supernatants very carefully with a P1000 pipet. Add 250 µL 70% ethanol (−20°C) to each tube, spin at 13,200g in a microcentrifuge at room temperature for 1 min. Remove supernatants very carefully. Air dry pellets completely.
Add 20 µL of sequencing gel loading buffer to each pellet. Incubate at 37°C for 20 min with constant mixing (see Note 11). Samples can be stored at −20°C.
Label some 10 bp DNA ladder as molecular weight standards for the sequencing gel (see Note 12).
Incubate sequencing gel samples and the labeled 10 bp DNA ladder at 65°C for 2 min and then rapid chill on ice. Load all 20 µL onto a 20% sequencing gel (see Note 13). Run until the bromophenol blue dye front has reached the bottom of the gel.
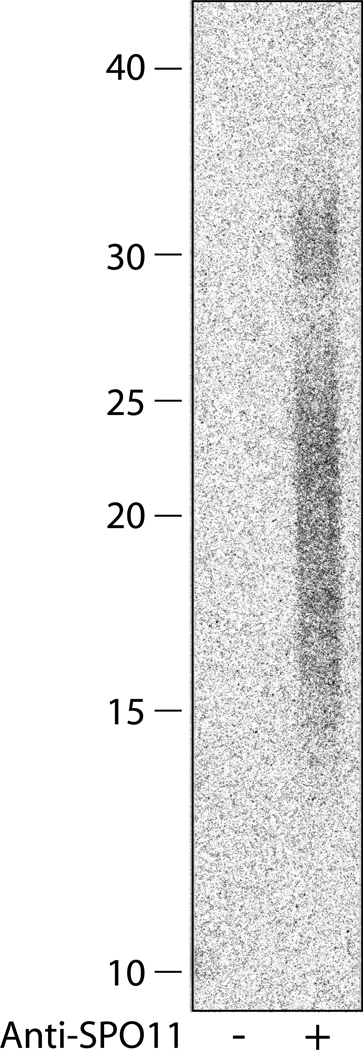
Incubate the gel in fixing solution for 10 min with mild agitation. Transfer the gel to water and incubate for another 10 min. Dry the gel onto a piece of cellulose paper backed by a piece of DE81 paper (see Note 13). Expose to a phosphor imaging plate overnight. SPO11-associated oligonucleotides from wild-type mouse testes span the region ~ 14–36 nt (Fig. 3).
Fig. 3.
Size of SPO11-associated oligonucleotides in wild-type mice. SPO11-associated oligonucleotide were labeled with [α-32P] cordycepin triphosphate and resolved on a 20% sequencing gel. Input to lanes: ~14 testes equivalent.
Footnotes
We use the same number of testes when comparing amounts of SPO11-oligonucleotide complexes among strains. Although SPO11-oligonucleotide complexes can be detected from two wild-type testes of an adult animal, we recommend using more material for robust signals. Four testes of each strain to be tested are used in this protocol for detection of SPO11-oligonucleotide complexes on an SDS gel, and experiments can be scaled up accordingly. If the size of SPO11-associated oligonucleotides is to be determined (Subheading 3.6.), at least eight testes of each strain should be used.
Fast, up-and-down movement is most effective to disrupt tissue and lyse cells using a plastic pestle. Take care to avoid spilling over during the grinding. As an alternative to plastic pestles, glass tissue grinders of a proper size can be used.
Do not take up supernatants by pipetting, because pipetting disturbs the pellets and causes a high background in the TdT labeling reaction. When the volume of the crude lysates is more than 2 mL, centrifugation is more conveniently carried out using either 3.5 mL or 5.5 mL-capacity ultracentrifuge tubes in a Sorvall AH-650 rotor (or equivalent) at 48,000 rpm (235,687g) for 30 min.
If precipitates appear after the incubation, spin briefly in a table-top centrifuge. Use supernatants for the following steps.
A slurry of protein A beads is made up with beads and buffer in approximately equal volume. To ensure that an equal amount of beads is added to each reaction, dispense aliquots of slurry into 1.5 mL tubes first. Spin beads down and make adjustment if there is an obvious difference in the volume of beads among tubes. Transfer the beads to immunoprecipitation reactions by resuspending the beads with 200 µL lysate.
We have also tested [α-32P]-labeled dGTP, dATP and the chain terminator cordycepin triphosphate in the TdT labeling reaction. There is no obvious difference in the migration of mouse SPO11-oligonucleotide complexes labeled by dCTP, dGTP or cordycepin triphosphate on the 8% SDS gel, and any of these nucleotides can be used in the labeling reaction. Do not use [α-32P] dATP because labeled SPO11-oligonucleotide complexes migrate slower and become less compact due to long and heterogeneous dA tailing. To determine the exact size of SPO11-associated oligonucleotides as described in Subheading 3.6., use cordycepin triphosphate in the labeling reaction.
Alternatively, the gel can be dried onto a piece of cellulose paper backed by a piece of DE81 paper. The dried gel is then wrapped and exposed to the imaging plate as described in Subheading 3.4., steps 4 and 5.
We observed different background labeling associated with TdT from different commercial sources, likely due to impurities in the TdT preparation. We recommend using FPLCpure TdT from GE Healthcare or TdT from New England Biolabs for this highly sensitive assay. A background smear at around 55 kD is present in the mock reaction when TdT from GE Healthcare is used in the labeling reaction (Fig. 1A). Two non-specific bands (about 75 kD and 100 kD) are present in the mock reaction when TdT from New England Biolabs is used (Fig. 1B).
We occasionally observe high lane background due to TdT labeling of contaminating DNA that is present in the immunoprecipitates (Fig. 1). Nevertheless, minor adjustment of brightness and contrast of the original image should reveal clear, specific signals of SPO11-oligonucleotide complexes. If SPO11-oligonucleotide complexes do not stand above background even after image adjustment, try repeating the experiment with more starting material. Alternatively, a second round of immunoprecipitation can reduce contaminating DNA significantly. Briefly, after the first round of immunoprecipitation (Subheading 3.2), elute the SPO11 by boiling the protein A beads in an equal volume of 2× SDS buffer. Save the SDS supernatant. Rinse the beads with 40 vol of wash buffer. Combine the washes with the SDS supernatant: this is eluate from the first immunoprecipitation. Perform the anti-SPO11 immunoprecipitation with the eluate, followed by TdT labeling, as described in Subheadings 3.2 and 3.3.
Tubes are fixed with tapes on the carousel in a hybridization oven and rotated slowly. Adjust the angle of the tubes relative to the rotating axis to make sure that the membrane has full contact with solution during the elution.
Tap the tubes to let the drop of liquid cover the rim above the cone bottom of the 2 mL tube, because much of the precipitate accumulates around the rim.
| 10 bp ladder (1 µg/ µL) | 1 µL |
| 10× T4 PNK buffer | 1 µL |
| [γ-32P] ATP | 2 µL |
| T4 PNK | 1 µL |
| 0.1 M DTT | 0.4 µL |
| Water | 4.6 µL |
| Total volume | 10 µL |
We run 27 cm long, 0.4 mm think sequencing gels with 6 mm wide wells using a custom-made apparatus. For detailed procedure on how to make, run, fix and dry a sequencing gel, please refer to Molecular Cloning (7).
References
- 1.Bellve AR. Purification, culture, and fractionation of spermatogenic cells. Methods Enzymol. 1993;225:84–113. doi: 10.1016/0076-6879(93)25009-q. [DOI] [PubMed] [Google Scholar]
- 2.Neale MJ, Pan J, Keeney S. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature. 2005;436:1053–1057. doi: 10.1038/nature03872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baudat F, de Massy B. Regulating double-stranded DNA break repair towards crossover or non-crossover during mammalian meiosis. Chromosome Res. 2005;15:565–577. doi: 10.1007/s10577-007-1140-3. [DOI] [PubMed] [Google Scholar]
- 4.Keeney S, Baudat F, Angeles M, Zhou ZH, Copeland NG, Jenkins NA, Manova K, Jasin M. A mouse homolog of the Saccharomyces cerevisiae meiotic recombination DNA transesterase Spo11p. Genomics. 1999;61:170–182. doi: 10.1006/geno.1999.5956. [DOI] [PubMed] [Google Scholar]
- 5.Romanienko PJ, Camerini-Otero RD. Cloning, characterization, and localization of mouse and human SPO11. Genomics. 1999;61:156–169. doi: 10.1006/geno.1999.5955. [DOI] [PubMed] [Google Scholar]
- 6.Sambrook J, Russel DW. Molecular Cloning. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2001. Chapter 12. Protocols 8, 11, and 12. [Google Scholar]