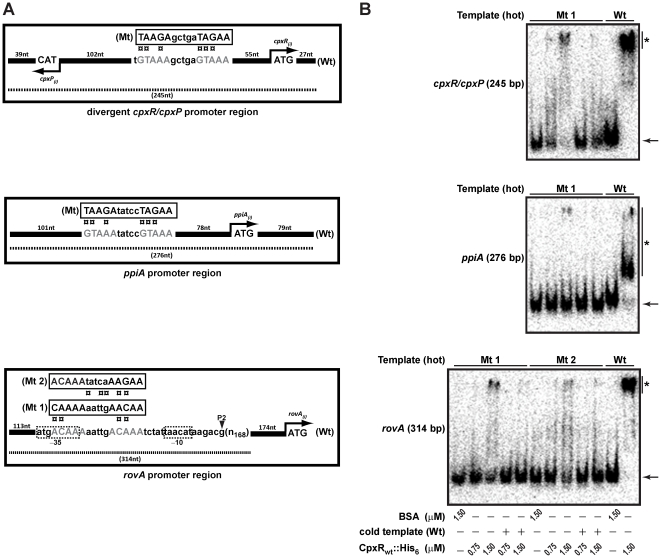
Figure 5. Site-directed mutagenesis of the CpxR∼P DNA binding site upstream of rovA.
(A) As identified by DNase 1 foot-printing (see Figure 4), the potential CpxR∼P binding site sequence and position is shown relative to each ATG start codon within the regulatory regions of rovA and ppiA and the divergent cpxR/cpxP promoter (designated as ‘Wt’). Site-directed mutagenesis was performed to ‘shuffle’ the nucleotide sequence of each potential CpxR∼P binding site (designated as ‘Mt’). Since the initial mutation in the rovA sequence (‘Mt 1’) would potentially disrupt RNA polymerase binding because of an altered −35 region within promoter P2, a second mutation (‘Mt 2’) was performed in which the −35 region was left untouched. (B) Mobility shift assays with purified CpxRwt::His6 as outlined in the legend to Figure 3 were performed on radiolabled amplified DNA from these mutated templates. Once again, the electrophoretic mobility of the ‘hot’ DNA fragments in the absence of bound protein is highlighted by an arrow, while DNA-CpxR∼P complexes are signified with a single asterisk (*).