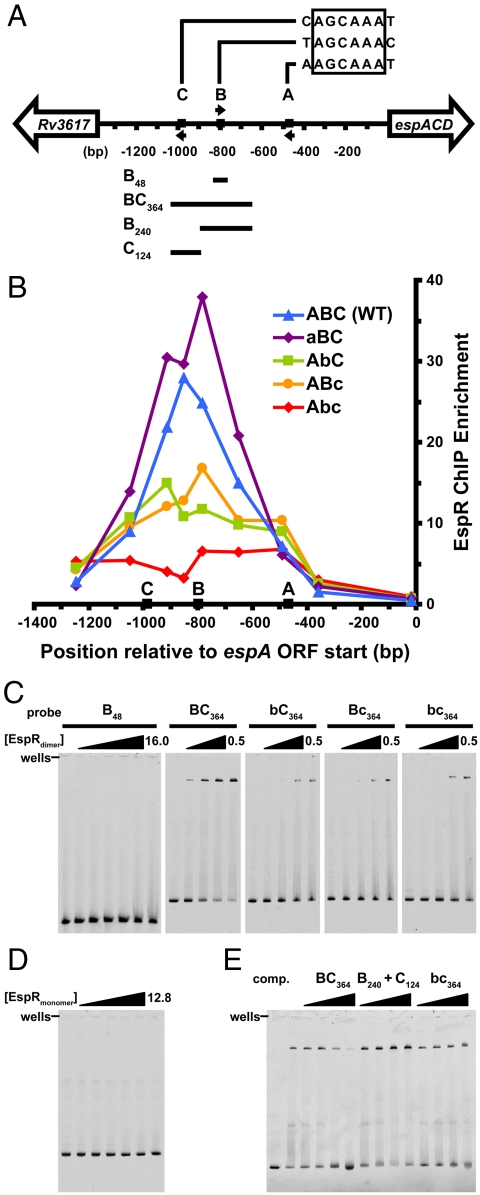
Fig. 4.
EspR binds far upstream of the espACD promoter and contacts two half-sites separated by 177 bp. (A) Map of the intergenic region upstream of the M. tuberculosis espACD operon, with EspR binding motifs A, B, and C, respectively, located at -468, -798, and -983 bp relative to the start of the ORF. Arrows indicate the orientation of each motif, and the four DNA fragments used for EMSA experiments are indicated below. (B) The wild-type intergenic sequence (ABC, blue triangles), and the same sequence containing scrambled-site mutations in A (aBC, purple diamonds), B (AbC, green squares), C (ABc, orange circles), and B and C together (Abc, red diamonds) were integrated into the M. smegmatis genome. M. tuberculosis EspR-3X-FLAG was expressed in each strain and ChIP-qPCR (quantitative PCR) was performed using anti-FLAG antibodies. Points represent the mean of triplicate qPCR measurements. (C) EMSAs were performed using purified EspR and the indicated 5-FAM-labeled probes (5 nM). For the B48 probe, EspRdimer concentrations were 0, 0.5, 1, 2, 4, 8, 16 µM, and for the BC364 probe and mutants, concentrations were 0, 0.2, 0.3, 0.4, and 0.5 µM. (D) The monomeric form of EspR (amino acids 1–107) was used in EMSAs with the BC364 probe ([EspRmonomer] = 0, 0.4, 0.8, 1.6, 3.2, 6.4, and 12.8 µM). (E) Binding for labeled BC364 probe ([EspRdimer] = 0.3 μM) was competed by adding unlabeled probes in 2-, 8-, 32-, and 128-fold molar excess of the labeled probe.