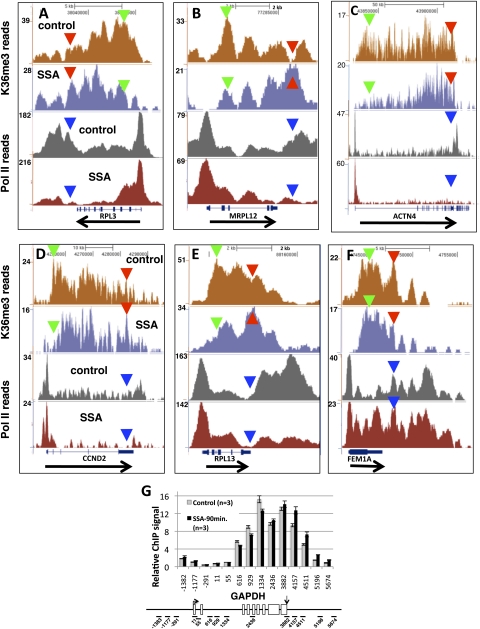
Fig. 3.
Inhibition of splicing causes widespread redistribution of histone H3K36me3 toward 3′ ends. Genome browser views of H3K36me3 and total pol II ChIP-seq reads in control HeLa cells and after treatment (20 ng/mL, 12 h) with the splicing inhibitor SSA for five intron-containing genes (A–E) and an intronless gene (F). Green and red or blue arrowheads mark 5′ and 3′ positions, respectively, for comparison of signals in WT and SSA. Note that the apparent 5′ to 3′ shift in H3K36me3 density is not paralleled by a similar shift in pol II density (see also SI Appendix, Fig. S4). (G) Relative H3K36me3 ChIP signals for 12 amplicons spanning the GAPDH gene in control HeLa cells (methanol treated) and after treatment with SSA (20 ng/mL) for 90 min. Relative ChIP signals were normalized to input and to amplicon +55 near the TSS. Means of 3 PCRs and SEMs are shown. Arrows mark the TSS and poly(A) sites in the map. Note the 3′ shift in H3K36me3 positioning similar to that detected in the 12-h treatment.