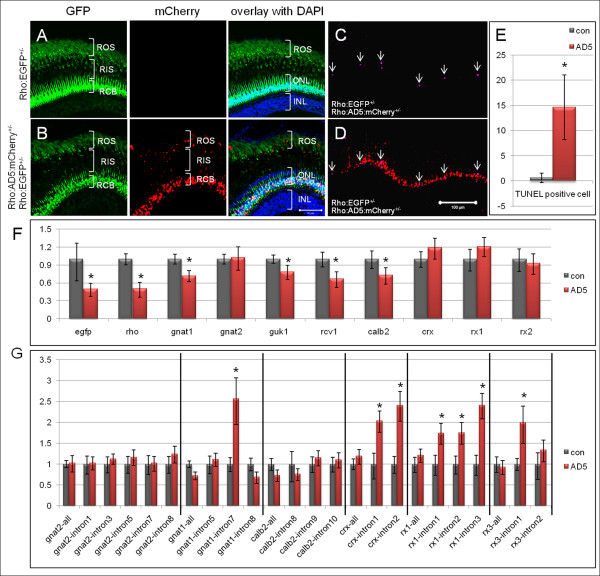
Figure 7.
Rod degeneration and splicing defects detected in adult AD5 transgenic fish. (A-E) Rod degeneration in adult AD5 transgenic fish. (A) Adult retina of Tg(Rho:EGFP) control fish at 5 months. EGFP is found in cell bodies (RCB) and outer segments (ROS), and can be seen faintly in inner segments (RIS). (B) Double transgenic fish expressing AD5:mCherry show multiple mCherry positive dots in the rod inner and outer segments, in addition to nuclear mCherry. (C) TUNEL positive apoptotic cells (arrow) in double transgenic fish expressing AD5:mCherry. (D) Overlay of TUNEL signal in image C with AD5:mCherry signal. (E) Number of TUNEL positive cells in double transgenic fish at 5 months expressing AD5:mCherry and Tg(Rho:EGFP) and in control fish (Tg(Rho:EGFP)). (F) qRT-PCR analysis showing gene expression profiles in double transgenic fish retinas expressing AD5:mCherry. 5 months old Tg(Rho:EGFP) transgenic fish retinas were used as control. Transcript numbers of several genes involved in photo-transduction are reduced, including rho, gnat1, guk1, recoverin, and calb2. egfp expression is also reduced, consistent with our histological findings. The cone specific transducin (gnat2) and several retina specific transcription factors (crx, rx1 and rx3) show no significant change. (G) Detection of splicing defects in double transgenic fish retinas (Tg(Rho:EGFP × Rho:AD5:mCherry)) by qRT-PCR. Among the six analyzed transcripts, introns from four genes showed significant stabilization compared to the same introns in control fish (Tg(Rho:EGFP), which includes intron 7 of gnat1, intron 1 and intron 2 of crx, intron 1, intron 2 and intron 3 of rx1, and intron 1 of rx3. In contrast, intron levels for cone specific gene gnat2 showed no significant change when compared to the control. Data were analyzed by T-test. Significant differences are indicated by asterisks. ONL, outer nuclear layer; INL, inner nuclear layer.