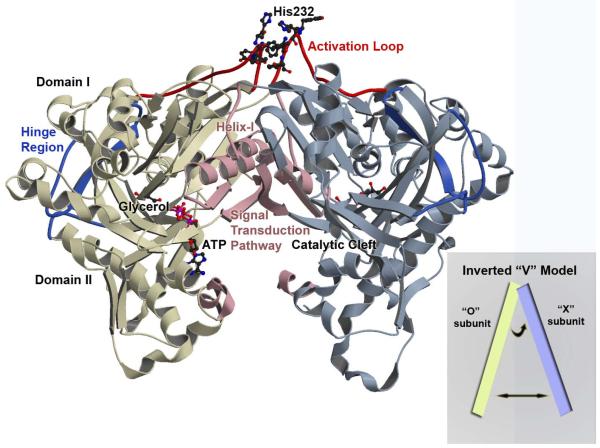
Figure 1. Overview of the Enterococcus casseliflavus GlpK O-X Dimer.
A ribbon depiction through Cα-backbone positions showing the overall topology and fold of the wild-type GlpK with the O-subunit colored in yellow and X-subunit in blue. Each subunit is composed of domain I and domain II with the catalytic cleft located at the interface of the two domains. The hinge region is colored in deep blue and delineates the boundary of the two domains and the location of the substrate binding site, with a molecule of glycerol depicted in ball-in-stick. The ATP-binding site is at the entrance of the catalytic cleft and ATP is modeled in ball-in-stick; its position is based on the E. coli ADP-complexed structure (PDB accession number 1GLB; 14) with the γ-phosphate position defined by the phosphate and sulfate ion location from the His232Arg and His232Glu GlpK structures, respectively. The activation loop is colored in red, with His232 and adjacent aromatic residues shown in ball-and-stick. Structural changes initiated by phosphorylation of His232 is transduced to the catalytic cleft along a pathway that is established by helix-I and the O-X dimer interface, colored in pink. The inset shows a schematic of the “inverted-V” topology, described in the text.