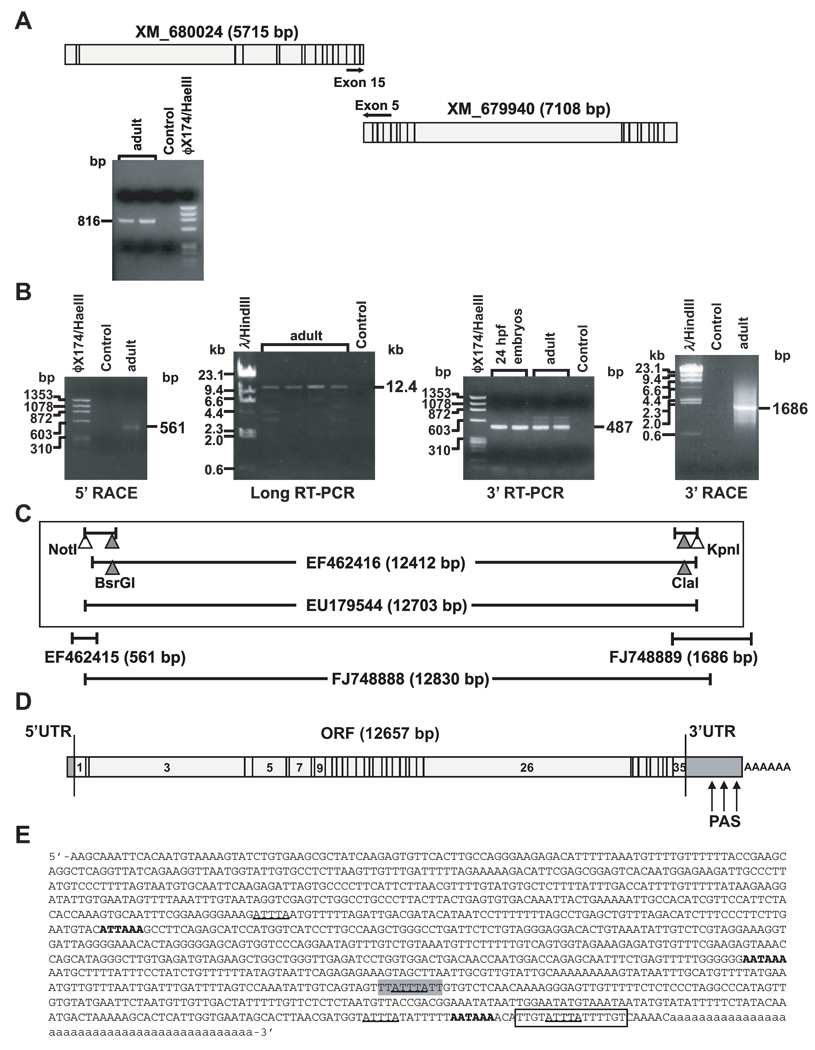
Fig 3.
Molecular cloning of zebrafish mll cDNA. (A) Reverse transcriptase (RT)-PCR design and products identifying single transcript spanning predicted cDNAs for both “similar to MLL proteins” in wild-type adult zebrafish total RNA. (B) Products of 5’ RACE containing 5’UTR (left), central ORF fragment obtained by long-range RT-PCR with gene-specific primers from predicted exons 1 and 35 (middle left, Accession no. EF462416), products of RT-PCR with primers from exon 33 and the predicted exon 35 containing the 3’ end of ORF to stop codon (middle right) and products of 3’ RACE PCR (right). (C) Strategy for subcloning cDNA spanning flanking sequence in 5’UTR and entire ORF through stop codon (Accession no. EU179544) into pCR-XL-TOPO vector (Top, boxed). Restriction sites in overlapping partial cDNAs (filled triangles) and engineered restriction sites (open triangles) that were used are indicated. cDNA subclones of 5’UTR (Accession no. EF462415), or spanning 3’ORF, 3’UTR and poly-A tail (Accession no. FJ748889) (middle), and cDNA subclone of entire ORF and partial flanking UTRs from single long-range PCR (Accession no. FJ748888) (bottom), are diagramed with respective GenBank entries. (D) Structure of zebrafish mll cDNA including UTRs. PAS indicates polyadenylation signals. (E) Sequence of zebrafish mll 3’UTR. AUUUA pentamers, underlined; UUAUUUAUU nonamer, grey highlight; polyadenylation signals corresponding to (D), bold; U-rich element, boxed; poly-A tail, lowercase.