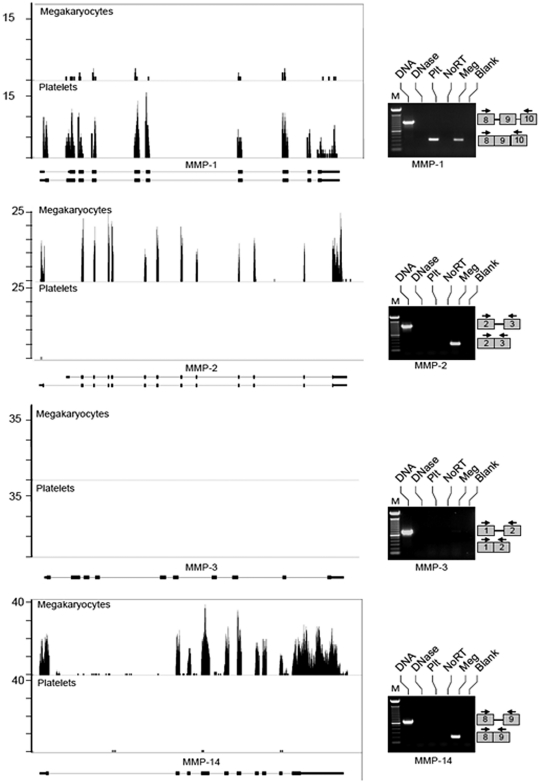
Figure 1.
Megakaryocytes and platelets differentially express MMP transcripts. mRNA was collected from megakaryocytes at the stage of proplatelet formation or freshly isolated, unactivated platelets and processed as described in “mRNA expression.” The left side of the figure shows RNA-seq analyses for MMP-1, MMP-2, MMP-3, and MMP-14 mRNA. Matched sequences are aligned to each target gene using the Integrated Genome Browser (IGB). Gene maps (bottom portion of each panel, oriented 5′-3′ direction) are represented by thick (exons) and thin (introns) lines. The bars above the gene (top portion of each panel) represent megakaryocyte or platelet transcripts that were fragmented, sequenced, and aligned to MMP-1, MMP-2, MMP-3, and MMP-14. If no bars are shown, then transcripts for these mRNAs were not detected under the conditions of these experiments. The right side of the figure shows semi-quantitative PCR analyses of mRNAs for MMPs 1-3 and MMP-14. From left to right, each lane represents: (1) amplification of DNA, which served as a positive control for primer efficiency; (2) DNA sample treated with DNase; (3) amplification of cDNA that was reverse transcribed from platelet (Plt) mRNA; (4) control reaction in platelet preparations that screened for residual genomic DNA contamination. In these studies, the reverse-transcription step was not performed (NoRT). (5) Amplification of cDNA that was reverse transcribed from megakaryocyte (Meg) mRNA; (6) negative control lane where the PCR was carried out without cDNA template (Blank). The PCR gels are representative of 5 or more independent experiments. The boxes on the right of each panel represent where the amplicons will migrate based on primer sequences specific for each exon.