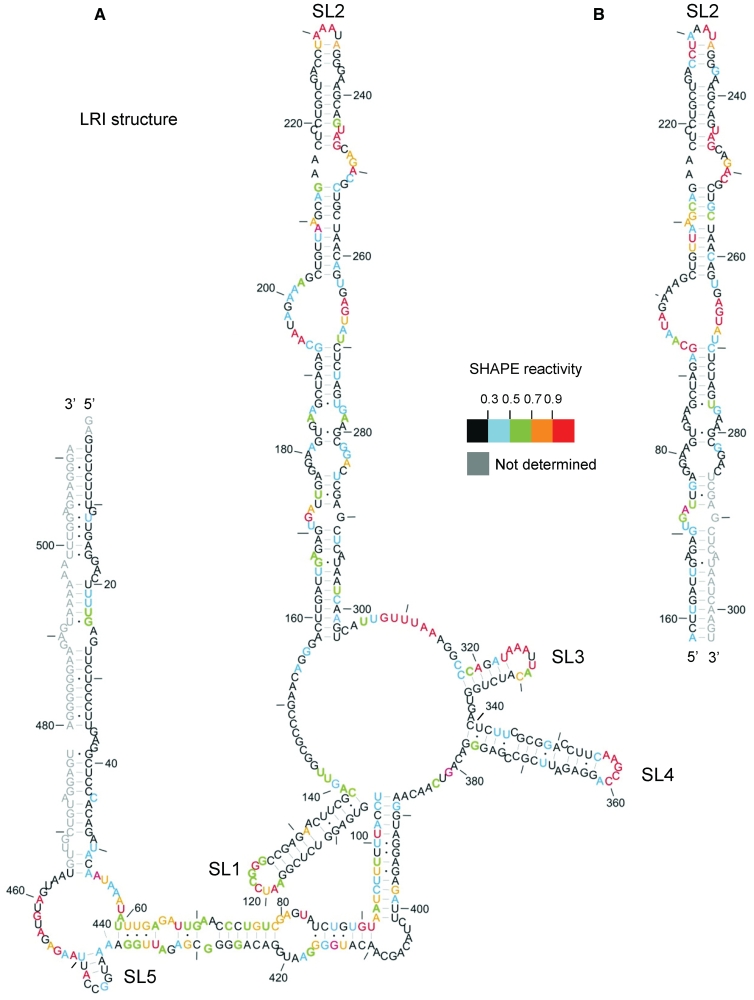
Figure 1.
SHAPE analysis of the FIV packaging signal RNA. In vitro transcribed RNA was modified with 1M7, reverse transcribed using fluorophore-labelled primers and separated by capillary electrophoresis. Nucleotide positions were determined using G and U sequencing ladders. 1M7 reactivity at each nucleotide position was determined by subtraction of the reverse transcription product of unmodified RNA from that of 1M7-modified RNA, using SHAPEfinder software. Nucleotide reactivities are colour-coded as shown in the key. Reactivities of the 3′-end were not determined (shown in grey). The RNA is drawn in our previously published secondary structure, to ascertain its validity. Nts are numbered with a dash every 10 nt. (A) In vitro transcribed packaging signal RNA; 511 nt. (B) In vitro transcribed SL2 RNA. Data shown are an average of at least two independent experiments.