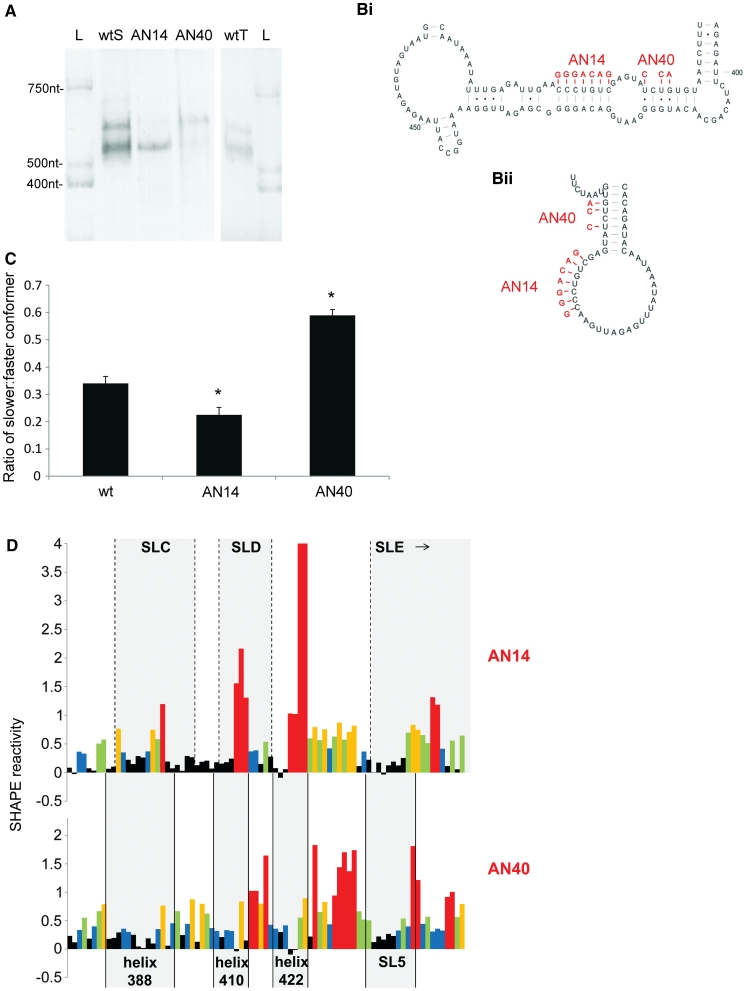
Figure 3.
Analyses of the two conformers by non-denaturing PAGE and SHAPE. (A) FIV packaging signal RNA was electrophoresed on a non-denaturing polyacrylamide gel and visualised by ethidium bromide staining and UV illumination. L; RNA ladder wtS; denatured and renatured wt RNA. AN14; denatured and renatured AN14 RNA. AN40; denatured and renatured AN40 RNA. wtT; wt RNA probed after transcription and column purification. (B) Schematic diagram showing the effects of AN14 and AN40 mutations upon: (Bi) the LRI structure; (Bii) SLB of the MSL structure. (C) Densitometric analysis of the ratio of slower-migrating to faster-migrating conformer. Data represent an average of at least six independent experiments. Stars represent statistical significance relative to wt (P < 0.01) by t-test. Error bars show the S.E.M. (D) NMIA reactivity profile of the 3′ portion of each mutant RNA from nts 380–460. Upper panel, AN14; lower panel, AN40. Grey boxes indicate the location of various structural features of each mutant. Reactivity >4 is shown as 4. Numbers below the graph represent the number of the initial nucleotide of each helix in the LRI structure. Data shown are an average of three independent experiments.