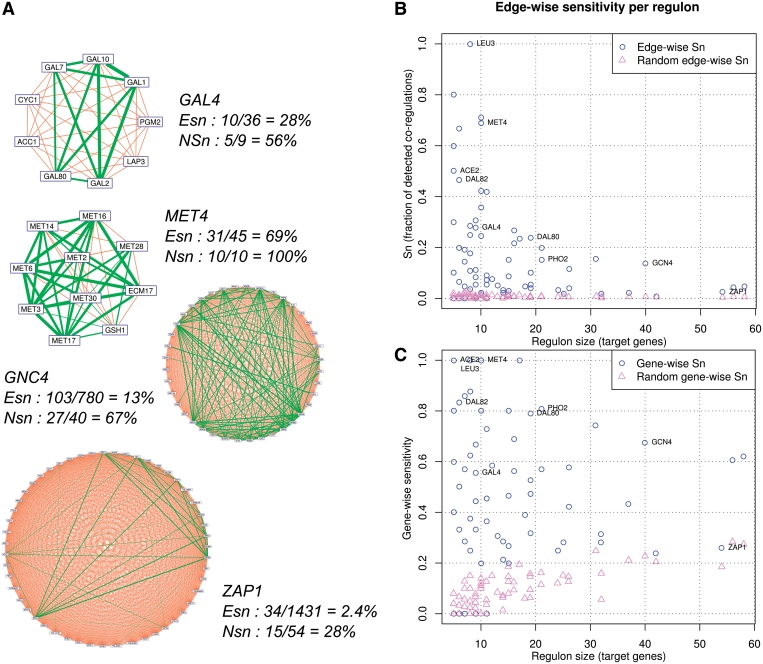
Figure 5.
Impact of regulon size on sensitivity. (A) Gene–gene cliques generated by specific (GAL4, MET4) and global (GCN4, ZAP1) TFs. Correctly predicted co-regulations (true positives) are highlighted in green, with edge thickness proportional to the DPbits score. Orange edges indicate false negatives (FN), i.e. existing co-regulations missing from the predicted co-regulation network. Edge-wise sensitivity (Esn) is the fraction of correctly predicted edges. Node-wise sensitivity (Nsn) is the fraction of target genes linked by at least one prediction. (B, C) Impact of regulon size (abscissa) on edge-wise (b) and gene-wise (c) sensitivity, respectively. All numerical values are provided in Table 4.