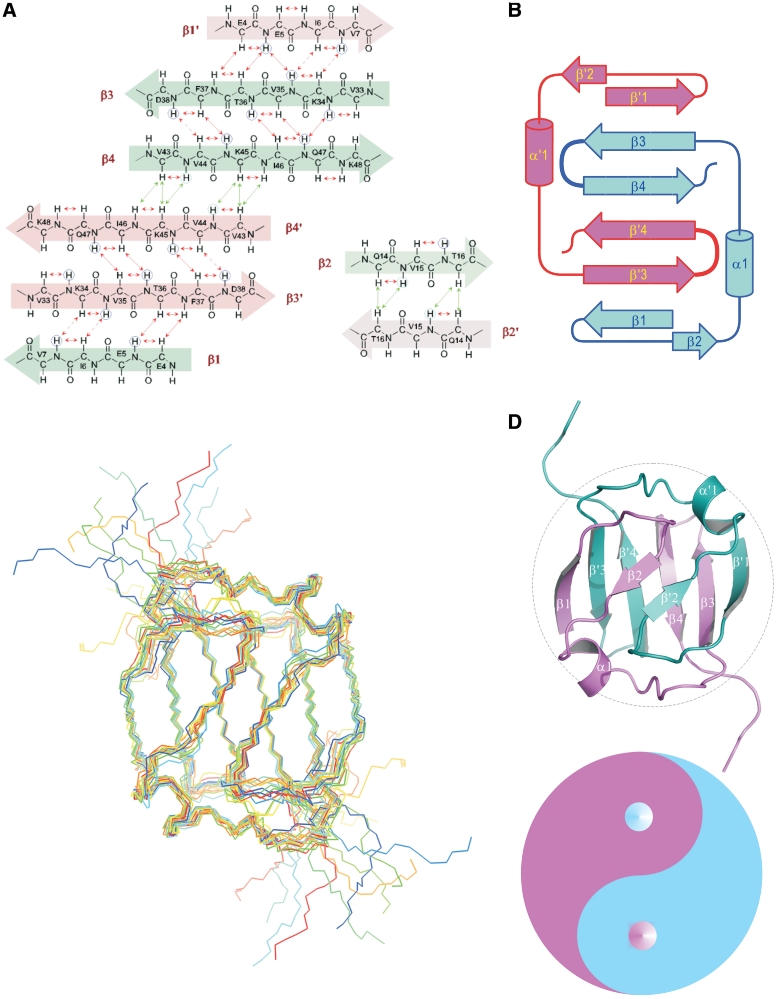
Figure 3.
Topology and NMR structure of the dimeric Sso7c4 protein. (A) The proton nuclear Overhauser enhancement (NOE) networks of the swapped six-stranded and the short two-stranded antiparallel β-sheets of Sso7c4 are defined from the NOEs and amide exchange rate. Long-range NOEs between β-strands are indicated by double arrows. The amide protons with very slow exchange rates are circled. (B) Topology diagram of the Sso7c4 structure showing the connectivity between strands in two β-sheets. (C) NMR ensemble of the selected structures is shown with a backbone chain. (D) The individual secondary structure elements are indicated in the ribbon diagram. The two monomers are colored differently for clarity. Association of the monomers is through two β-sheets; in each sheet, two strands derive from one monomer and the first strand forms the other monomer. In addition, another short β-sheet is constructed by the β2 strand from each monomer. These arrangements form a strand-switched dimer interface. The architecture of homodimeric Sso7c4 is presented as the Chinese traditional ‘Tai-Chi’ symbol. The separation line (also called ‘Yin-Yang’ diameter) formed by two semicircles of the ‘Tai-Chi’ symbol generate more binary interaction than does the linear diameter.