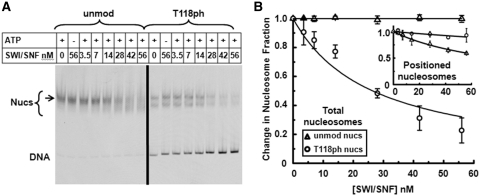
Figure 8.
H3(T118ph) enables nucleosome disassembly by the SWI/SNF remodeling complex. (A) EMSA of ATP-dependent chromatin remodeling with unmodified and H3(T118ph) nucleosomes in the presence of increasing concentrations of SWI/SNF. Lanes are labeled with the nanomolar (nM) concentration of SWI/SNF. Each reaction was incubated for 30 min with or without 1 mM ATP. The bracket indicates the range of nucleosome electrophoretic mobility and the arrow indicates the location of central positioned nucleosomes. (B) The change in the fraction of total unmodified (triangles) or H3(T118ph) (circles) nucleosomes versus SWI/SNF concentration. The error bars were determined from the standard deviation of three separate experiments. The total unmodified nucleosome fractions and the total H3(T118ph) nucleosome fractions were each fit to: nucleosome fraction = 1-[SWI/SNF]/(K½+[SWI/SNF], where K½ is the concentration at which half of the nucleosomes are disassembled. We find that there is not a decrease in the fraction of unmodified nucleosomes (K½, unmod total > 2 × 10−5 nM), while K½,H3(T118ph) total = 26 ± 4 nM. (B, inset) The change in the fraction of centrally positioned unmodified (triangles) and H3(T118ph) (circles) nucleosomes versus SWI/SNF concentration. We fit the faction of positioned nucleosomes to the equation used to fit the total nucleosome fraction. We find H3(T118ph) decreases by 6-fold the fraction of positioned nucleosomes (K½, unmod positioned = 80 ± 3 nM and K½,H3(T118ph) positioned = 500 ± 100 nM). The values were determined from the ratio of the top band (arrow) to the entire nucleosome band (bracket) and normalized by the fraction of positioned nucleosomes at zero concentration of SWI/SNF.