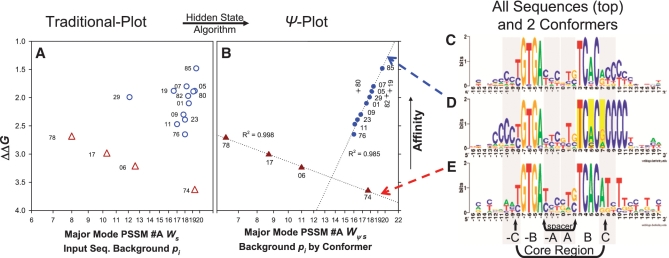
Figure 4.
Affinity (ΔΔG) versus weight score (Ws) comparison validating the HS model. Traditional (open symbols) and Ψ-plots (closed symbols) for dyad (triangles) and dyad (+10)bend (circles and crosses) EcCrp/XD-DNA sequence-specific conformers. Substrate ΔΔG values relative to ΔG for ICAP were obtained from experiments performed by Lindemose et al. (34). PSSM #A was the scanning matrix for calculating all scores. The highest score fitting to PSSM #A (either strand orientation) is shown. (A) PSSM #A calculated Ws of dyad (open triangles) and dyad (+10)bend substrates (open circles). (B) PSSM #A calculated ψ-scores (Wψs) of ψEcCrp/XD-DNA for dyad (closed triangles) and dyad (+10)bend substrates (closed circles and crosses). Each set of conformer-specific sequences scored (e.g. the circles) make up the input sequence background pi for scoring that conformer. Specific ψ-sequences, ΔΔG values, and PSSM #A calculated ψ-scores are clearly listed in Supplementary Table S36. Outliers (crosses) are labeled vertically and not included in the R2 value because they are distinct from a trendline. R2 shown for trend lines (dotted) was obtained by fitting to y = y0 + anlnx. ΔΔG is the same for any given substrate in each graph. The point for the highest affinity substrate is G8.85 = 85 and labels correspond to the G8.## clones as described (34). Note logarithmic scaling of the abscissa. For the XD-DNA shown, G = X and A = D. (C) The entire sequence distribution constituting the major binding mode illustrated as PSSM #A (25.7 bits). All 49 unique sequences of the SELEX system make up this logo. The EcCrp/XD-DNA complex affinities for 26 dsDNA substrates with these unique sequences were determined, leaving 23 sequences having unknown affinities. (D) The dyad (+10)bend conformer illustrated as PSSM #A1 (33.5 bits). The 14 sequences that contain (+10)CCCC make up this logo. A total of 12 affinities were determined (circles and crosses in A and B). (E) The dyad conformer illustrated as PSSM #A2 (31.2 bits). The seven sequences that contain ≤2 X/C bp's total in both (+ and −10)NNNN tracts make up this logo. A total of four affinities were determined (triangles in panels A and B). The reported bit values span between positions −16 and +16. The logos in C and D are not ψ-sequences. Base positions and arbitrary regions shaded in gray are labeled as in Figure 1A. Flanking flexible bend proximal primary kink associated positions +3, +5 and +7 (yellow bars) are where ψ-sequence changes were performed.