Abstract
Entamoeba histolytica is the causative agent of amebiasis and infects up to 10% of the world's population. The molecular techniques that have enabled the up- and down-regulation of gene expression rely on the transfection of stably maintained plasmids. While these have increased our understanding of Entamoeba virulence factors, the capacity to integrate exogenous DNA into genome, which would allow reverse genetics experiments, would be a significant advantage in the study of this parasite. The challenges presented by this organism include inability to select for homologous recombination events and difficulty to cure episomal plasmid DNA from transfected trophozoites. The later results in a high background of exogenous DNA, a major problem in the identification of trophozoites in which a bona fide genomic integration event has occurred. We report the development of a negative selection system based upon transgenic expression of a yeast cytosine deaminase and uracil phosphoribosyl transferase chimera (FCU1) and selection with prodrug 5-fluorocytosine (5-FC). The FCU1 enzyme converts non-toxic 5-FC into toxic 5-fluorouracil and 5-fluorouridine-5'-monophosphate. E. histolytica lines expressing FCU1 were found to be 30 fold more sensitive to the prodrug compared to the control strain.
Protocol
1. FCU Negative Selection System in Entamoeba histolytica
The vector control line (HM1/ pKT3M) and the experimental line (HM1/pKT3M-FCU1) were generated by previously described techniques1,2. Seed the lines under study from the stock tubes into T-25 flasks and maintain selection by adding appropriate antibiotic (12 μg/ mL G418) in TYI-S-333 medium. The flasks should be 70-80% confluent after 16-18 hours of growth at 37°C.
Prepare a fresh 50mM stock solution of 5-Fluorocytosine (5-FC, Sigma) in warm TYI-S-33 medium. Vortex rigorously for several minutes to dissolve 5-FC completely. Filter-sterilize the stock solution by passing it through a 0.22 μm filter.
Harvest trophozoites by placing the flasks on ice for 3-5 minutes and collecting the cells by centrifuging at 200xg for 5 minutes at room temperature. Resuspend the pellet in fresh TYI medium and count the cells.
Design of a typical experiment uses 0.5 x 104 cells per well of a 48 well plate in a final volume of 1 mL; and triplicate repeats for each test condition. In addition, to facilitate fluorescent measurements, a separate plate is used for each strain per time point.
Seed 0.5 x 104 cells per well for each test 5-FC concentration, in triplicate. Maintain the antibiotic selection pressure in the TYI medium. Now add the required amount of 5-FC stock solution and trophozoites to each well.
Incubate the plates in an anaerobic bag at 37°C.
2. Determination of Selection Efficiency
Prepare 2 mg/ mL stock solution of Cell tracker green CMFDA (Invitrogen) in DMSO. To make the working solution dilute the stock 1000-fold in serum-free TYI medium.
Remove the medium completely from trophozoite culture plates and replace with 150 μL serum-free TYI containing CMFDA. Continue incubation at 37°C for 1 hr.
Remove the dye solution from wells and read the plate in the Spectramax M2 microplate spectrofluorometer. The excitation wavelength should be set at 492 nm and the fluorescence emission read at 517 nm.
Compare the fluorescence values for the control cells versus the test transfectants.
Continue reading the plates at each time interval. Use appropriate positive (TYI only) and negative (25 μg/ mL hygromycin) control wells in each plate.
3. Representative Results
The E. histolytica transfectants showed robust expression of codon optimized recombinant FCU1 (Figure 1).When this protocol is followed correctly it results in selective elimination of E. histolytica cells expressing FCU1 in presence of 0.5 mM 5-fluorocytosine. Control cells, in contrast continue to grow normally at up to 5 mM concentration of 5-FC and reach confluence between 72 and 96 hrs (Figure 2-a). The FCU1 carrying cells at 48 hrs are completely lysed when the treated wells are viewed under a microscope. These also show decreased fluorescence when stained with the vital dye CMFDA, which is used in quantitative studies involving large number of samples (figure 2-b). The FCU1/5-FC system is an effective and powerful negative selection tool for E. histolytica trophozoites.
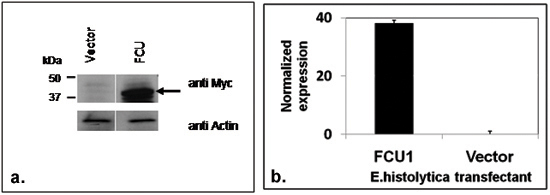 Figure 1. Generation of E. histolytica HM1 transfectants expressing negative selectable marker
(a) FCU1 recombinant protein was detected on a western blot using anti-c Myc antibody (top panel). As expected, a 42kDa band could be detected (shown by an arrow) in the total lysate from the FCU transfectants but not in the vector alone control transfectants. The bottom panel shows the same blot probed with anti actin antibody as a loading control. (b) Results of the quantitative real time PCR showing expression of FCU1 specific mRNA normalized to lgl4 control.
Figure 1. Generation of E. histolytica HM1 transfectants expressing negative selectable marker
(a) FCU1 recombinant protein was detected on a western blot using anti-c Myc antibody (top panel). As expected, a 42kDa band could be detected (shown by an arrow) in the total lysate from the FCU transfectants but not in the vector alone control transfectants. The bottom panel shows the same blot probed with anti actin antibody as a loading control. (b) Results of the quantitative real time PCR showing expression of FCU1 specific mRNA normalized to lgl4 control.
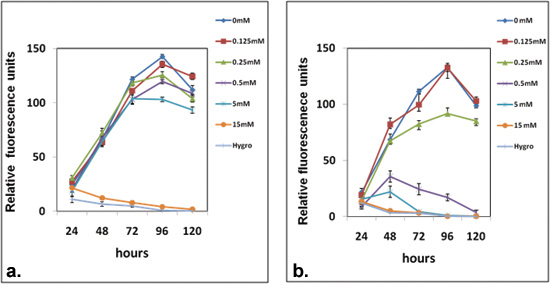 Figure 2. In vitro drug sensitivity of E. histolytica transfectants expressing negative selectable marker
Survival curves of (a) control transfectants and (b) FCU1 expressing transfectants cultured in 48-well plate. The cells were grown in presence of increasing concentrations of the prodrug- 5-fluorocytosine; cell survival was quantified by CMFDA staining at each time interval as denoted on the X-axis. The Y-axis represents fluorescence units and is a direct reflection of the surviving cell population. Please note that the FCU1 expressing transfectants showed significant sensitivity at 0.5mM prodrug concentration compared to the control. No cells were observed at 120h time point in these wells as compared to the confluent growth seen in the corresponding control wells under microscope (data not shown). Hygro denotes 25μg/ mL hygromycin used as a negative control.
Figure 2. In vitro drug sensitivity of E. histolytica transfectants expressing negative selectable marker
Survival curves of (a) control transfectants and (b) FCU1 expressing transfectants cultured in 48-well plate. The cells were grown in presence of increasing concentrations of the prodrug- 5-fluorocytosine; cell survival was quantified by CMFDA staining at each time interval as denoted on the X-axis. The Y-axis represents fluorescence units and is a direct reflection of the surviving cell population. Please note that the FCU1 expressing transfectants showed significant sensitivity at 0.5mM prodrug concentration compared to the control. No cells were observed at 120h time point in these wells as compared to the confluent growth seen in the corresponding control wells under microscope (data not shown). Hygro denotes 25μg/ mL hygromycin used as a negative control.
Discussion
Although there have been substantial advances in Entamoeba molecular biological tools there are no current methods available to delete or disrupt genes4. Knockdown of gene specific expression has been achieved by the use of hairpin RNAs5, small interfering RNA6 and bacterially expressed dsRNA7. However these methods while effective for the respective target genes, have several limitations including use of costly chemicals, continuous selection against antibiotics and the need for constant validation due to the high incidence of reversion occurring over time (Alicia Linford, personal communication)8.
Any plasmid can replicate in Entamoeba as there is apparently not a strict sequence requirement for origins of DNA replication9, and so once transfected, it usually is difficult to cure a plasmid. This limits the possible use of intact plasmids in recombination and gene knockout experiments. Negative selection markers are key components of gene targeting methods as they can be used to eliminate recombinant subpopulations. We have successfully expressed a codon optimized FCU1 gene in Entamoeba histolytica and tested its potential as a negative selectable marker. This gene fusion has been used for suicide gene therapy of human tumor cells and metabolizes the prodrug 5-FC into toxic downstream products resulting in inhibition of RNA and DNA synthesis10. FCU1 has been recently used as a negative selectable marker in Plasmodium, another protozoan parasite. Plasmodium berghei cells expressing FCU1 were found to be almost 1000 fold more sensitive to the prodrug in vitro and in vivo treatment with 5-FC killed over 99.9% of the infecting recombinant parasites in the erythrocytic-stage mouse model of malaria11. E. histolytica cells expressing FCU1 showed only a 30-fold increased sensitivity to the prodrug 5-FC in contrast to the sensitivity observed in P. berghei. Possible reasons for this could be large pool of nucleotides available in the growth medium competing with the toxic metabolites. In both P. berghei and P. falciparum the FCU1 marker has been used in targeted gene disruption. studies11,12. We aim to manipulate the Entamoeba genome using homologous recombination. The validation of the FCU1/5-FC negative selection system is a significant step towards this goal.
Disclosures
The production of this video was sponsored by Sigma Aldrich, Inc, which produces some of the reagents used in this article.
Acknowledgments
We would like to thank Dr. Philippe Erbs for providing us with the sequence of FCU1 gene. This work was supported by NIH grant AI 26649.
References
- Olvera A. Stable transfection of Entamoeba histolytica trophozoites by lipofection. Arch. Med. Res. 1997:49–51. [PubMed] [Google Scholar]
- Saito-Nakano Y, Yasuda T, Nakada-Tsukui K, Leippe M, Nozaki T. Rab5-associated vacuoles play a unique role in phagocytosis of the enteric protozoan parasite Entamoeba histolytica. J. Biol. Chem. 2004;279:49497–49507. doi: 10.1074/jbc.M403987200. [DOI] [PubMed] [Google Scholar]
- Diamond LS, Harlow DR, Cunnick CC. A new medium for the axenic cultivation of Entamoeba histolytica and other Entamoeba. Trans. R. Soc. Trop. Med. Hyg. 1978;72:431–432. doi: 10.1016/0035-9203(78)90144-x. [DOI] [PubMed] [Google Scholar]
- Clark C. Anaerobic parasitic protozoa : genomics and molecular biology. Norfolk UK: Caister Academic; 2010. [Google Scholar]
- Linford AS. Short hairpin RNA-mediated knockdown of protein expression in Entamoeba histolytica. BMC Microbiol. 2009;9:38–38. doi: 10.1186/1471-2180-9-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solis CF, Guillén N. Silencing genes by RNA interference in the protozoan parasite Entamoeba histolytica. Methods Mol. Biol. 2008;442:113–128. doi: 10.1007/978-1-59745-191-8_9. [DOI] [PubMed] [Google Scholar]
- Solis CF, Santi-Rocca J, Perdomo D, Weber C, Guillén N. Use of bacterially expressed dsRNA to downregulate Entamoeba histolytica gene expression. PLoS ONE. 2009;4:e8424–e8424. doi: 10.1371/journal.pone.0008424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacFarlane RC, Singh U. Loss of dsRNA-based gene silencing in Entamoeba histolytica: implications for approaches to genetic analysis. Exp. Parasitol. 2008;119:296–300. doi: 10.1016/j.exppara.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar SK, Vines RR, Bhattacharya S, Petri WA. Ribosomal DNA fragments enhance the stability of transfected DNA in Entamoeba histolytica. J. Eukaryot. Microbiol. 1998;45:656–660. doi: 10.1111/j.1550-7408.1998.tb04563.x. [DOI] [PubMed] [Google Scholar]
- Erbs P. In vivo cancer gene therapy by adenovirus-mediated transfer of a bifunctional yeast cytosine deaminase/uracil phosphoribosyltransferase fusion gene. Cancer Res. 2000;60:3813–3822. [PubMed] [Google Scholar]
- Braks JAM, Franke-Fayard B, Kroeze H, Janse CJ, Waters AP. Development and application of a positive-negative selectable marker system for use in reverse genetics in Plasmodium. Nucleic Acids Res. 2006;34:e39–e39. doi: 10.1093/nar/gnj033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier AG, Braks JAM, Waters AP, Cowman AF. Negative selection using yeast cytosine deaminase/uracil phosphoribosyl transferase in Plasmodium falciparum for targeted gene deletion by double crossover recombination. Mol. Biochem. Parasitol. 2006;150:118–121. doi: 10.1016/j.molbiopara.2006.06.014. [DOI] [PubMed] [Google Scholar]


