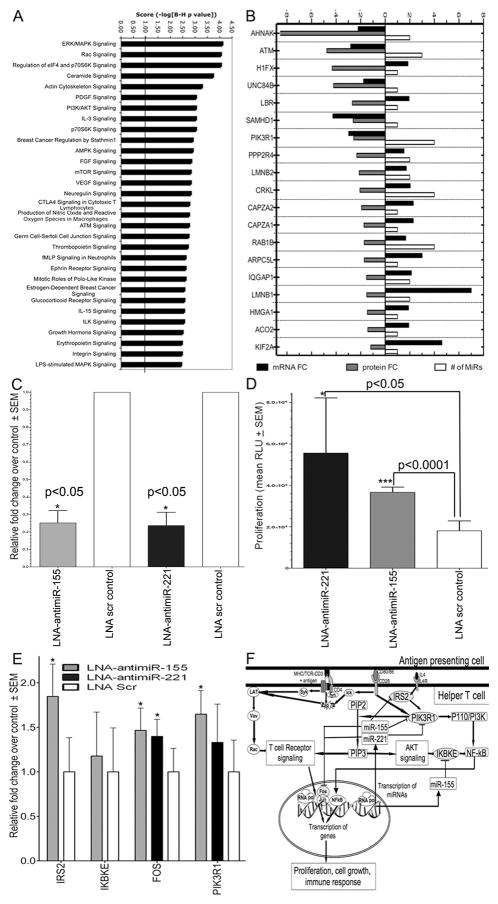
Figure 4.
Inhibition of miR-155 (known) and miR-221 (novel), 2 highly upregulated miRNAs, in CD4+ T cells increased proliferation by removing suppression of 4 target genes linked to proliferation and survival. (A) Top 30 overrepresented canonical pathways for downregulated protein targets of upregulated miRNAs. Pathways are sorted by score. (B) Expression of 19 predicted downregulated protein targets associated with top 3 networks. (C) Real-Time qPCR expression of miR-155 and miR-221 in CD4+ T cells nucleoporated with 50nM LNA-antimiR-155 (tan), LNA-antimiR-221 (black), or LNA-scrambled control showing decreased miRNA expression after LNA-antimiR transfection relative to scrambled control, set as 1. Shown are fold changes relative to scrambled control, normalized to U6 snRNA. (D) Increased cell proliferation following knockdown of miR-221 (black) or miR-155 (tan), compared to scrambled control by Cell Proliferation Assay. (E) Real-Time qPCR analysis of predicted targets in CD4+ T cells transfected with LNA-antimiR-155 (tan), LNA-antimiR-221 (black), and LNA scrambled control, presented relative to the expression in scrambled control sample, set as 1. (F) Proposed model of miR-155 and 221 negative-feedback regulation of proliferation following T cell activation. CD3/CD28 co-stimulation induces signaling cascades that result in transcription of genes that promote proliferation, activation and immune response. Upregulation of miR-155 and 221 serves to diminish proliferation and immune response by targeting key proliferative genes such as PIK3R1, IRS2 and IKBKE as well as transcription factor FOS. Error bars in (C–E) represent mean ± SEM of triplicate experiments from 3 donors (n=3). *p < 0.05, ***p < 0.0001 (t-test) compared to scrambled control.