Abstract
A methodology is proposed for nonlinear contrast-enhanced unsupervised segmentation of multispectral (color) microscopy images of principally unstained specimens. The methodology exploits spectral diversity and spatial sparseness to find anatomical differences between materials (cells, nuclei, and background) present in the image. It consists of rth-order rational variety mapping (RVM) followed by matrix/tensor factorization. Sparseness constraint implies duality between nonlinear unsupervised segmentation and multiclass pattern assignment problems. Classes not linearly separable in the original input space become separable with high probability in the higher-dimensional mapped space. Hence, RVM mapping has two advantages: it takes implicitly into account nonlinearities present in the image (ie, they are not required to be known) and it increases spectral diversity (ie, contrast) between materials, due to increased dimensionality of the mapped space. This is expected to improve performance of systems for automated classification and analysis of microscopic histopathological images. The methodology was validated using RVM of the second and third orders of the experimental multispectral microscopy images of unstained sciatic nerve fibers (nervus ischiadicus) and of unstained white pulp in the spleen tissue, compared with a manually defined ground truth labeled by two trained pathophysiologists. The methodology can also be useful for additional contrast enhancement of images of stained specimens.
Staining of the specimen in the slide preparation process has been standard procedure for many years, because it increases contrast between the cell and the background. However, staining involves hours of preprocessing of the specimen, and can also add chemical effects to the nature of the cells, cause their shrinkage, and alter their morphology.1 For example, in studying effects of DNA damage on cell viability, fluorescent probes must not be used to stain the cell nuclei, to avoid compromising the viability of the cultures.2 Similarly, when studying the effects of inhibitor compounds designed to block the replication of cancerous cells, fluorescent dyes must not be used to mark nuclei, because the dyes themselves have toxicity.3 The subcellular localization of genetically encoded proteins imposes constraints on the cell recognition methods used to draw conclusion about function of a protein; again, staining of the cell is not allowed, to preserve the quality of the specimen and to avoid influencing the result of an investigation.4 When staining is not allowed, contrast between the cell and the background will be poor, and it is challenging for either a trained pathologist or an automated image processing system to achieve good results. Furthermore, manual classification of cells by trained pathologists in images of stained specimens shows interobserver variability up to 20%.5 These findings prompted us to explore the possibility of developing a contrast enhancement methodology based on nonlinear unsupervised segmentation of multispectral images with poor contrast between the materials (ie, cells, nuclei, tissue types, and the like) present in the image.
The methodology reported here is based on representation of the multispectral image by a linear mixture model (LMM) in a space induced by rth-order rational variety mapping (RVM), which is a polynomial type of nonlinear mapping.6 LMM itself has been used in remote sensing for years,7–9 and recently in medical imaging as well.10–12 It represents a multispectral image matrix as a product of basis or mixing matrix and matrix of spatial distributions of the materials present in the image (referred to as a source matrix). Very recently, the tensorial nature of the multispectral image has been exploited for the purpose of linear unsupervised segmentation.13 Thus, any method capable of uniquely factorizing multispectral image matrix or tensor can be used to perform unsupervised segmentation. This implies that only a multispectral image is at disposal for the factorization method. In adopted representation, columns of the basis matrix represent spectral profiles of the materials present in the image.
The quality of linear segmentation depends in the first place on how accurately LMM represents an experimental multispectral image. When nonlinearities are present in the image, accuracy of the LMM-based representation is limited. It can, however, be improved if LMM is used to represent the image in the space induced by the rth-order RVM.6 This capability is grounded in the duality existing between nonlinear unsupervised image segmentation and pattern recognition problems, such as occurs when spatial distributions of the materials present in the image are mutually sparse. Here, RVM increases the number of separating surfaces between the classes and enables their separability. Then, the multispectral image represents a set of patterns (pixel vectors), columns of the mixing matrix represent centers of the class-related clusters, and sources represent classes. Representing the image by LMM in mapped space has two advantages: it implicitly takes into account the possible presence of nonlinearities in the image (ie, they are not required to be known) and it enhances contrast between spectrally similar materials, which occurs because of increased dimensionality of the mapped space. In this regard, nonlinear band expansion as used by Kopriva and Peršin10 and by Ouyang et al 14 is a special case (second-order) of the rth-order RVM used in our methodology.
We apply second- and third-order mappings (RVM2 and RVM3, respectively) in combination with several state-of-the-art matrix and tensor factorization methods15–18 to perform nonlinear unsupervised segmentation of the multispectral microscopy images of the unstained specimen of sciatic nerve fibers (nervus ischiadicus) and spleen tissue. In the first case, our proposed methods yielded significant contrast enhancement between nerve cells and background, relative to segmentation performed by factorization algorithms only. The same outcome occurred in the second experiment: with our proposed methods, lymphatic cells present in the white pulp spleen tissue were better discriminated against the red pulp, relative to segmentation performed by factorization algorithms only. Moreover, RVM3 brought an additional performance improvement. The proposed methodology can also be useful for additional contrast enhancement in analysis and classification problems associated with multispectral images of stained specimens.19–23 To our knowledge, there is no previous report of nonlinear blind segmentation of multispectral (color) images of either unstained or stained specimens performed by matrix/tensor factorization in the space induced by rth-order RVM.
Materials and Methods
Animal Studies
Adult NOD (10 to 12 weeks old) mice were acquired from the breeding facility at the Ruđer Bošković Institute, Zagreb, Croatia. Animal care and treatment were conducted in conformity with institutional guidelines and in compliance with international law. Before and during the experiments, animals had standard pelleted food and tap water ad libitum. The studies were approved by the institutional Ethics Committee.
Histological Analysis
Sciatic nerve fibers and spleen tissue were taken from NOD mice for histological analysis. After extirpation, the tissues were fixed in Bouin's solution for 24 hours. Fixed tissues samples were immersed in increasing concentrations of alcohol for dehydration. After dehydration, tissues were embedded in paraffin. The paraffin blocks were then cut with a microtome into slices 5 μm thick.
Deparaffinization
Microscope glass slides with a histological preparation were put in a rack, covered with foil, and incubated for 5 minutes at 55°C in an oven (Thermo Fisher Scientific, Waltham, MA). Afterwards, glass slides were incubated three times for 15 minutes in toluene. Microscope glass slides with a histological preparation were put in a rack, covered with foil, and incubated for 5 minutes at 55°C in an oven (Thermo Fisher Scientific, Waltham, MA). Afterwards, glass slides were incubated three times for 15 minutes in toluene. Glass slides were then transferred into 100% ethanol at 1 minute, after that started process rehydration for 5 minutes in 96% ethanol, for 5 minutes in 75% ethanol, and for two 5-minute washes in distilled water with constant stirring.
Preparation for Staining and Antigen Retrieval
Deparaffinized microscope glass slides with histological preparation were placed in a rack, plunged into 450 mL citrate buffer, and heated two times for 5 minutes in a microwave oven. Slides were then transferred into hot water, heated to 60°C, and stored until cooled down to room temperature. After cooling, slides were washed three times for 5 minutes in PBS buffer at room temperature.
Staining
The microscopic glass slides with histological preparation were applied to 50 μL incubation buffer. After incubation and mixing (for 30 minutes) glass slides were washed with PBS buffer. On each slide, 50 μL primary monoclonal antibody (mAb IgG) was added and slides were incubated for 60 minutes with occasional mixing. After incubation, the slides were rinsed thoroughly three times for 5 minutes in PBS buffer. Next, 80 μL 10% glycerol-PBS was added to each slide; slides were then coverslipped. Finally, processed glass slides were analyzed under a microscope.
Multispectral Microscope Image Acquisition
Spectral images of the slides were acquired at room temperature, in the 10% glycerol-PBS imaging medium, under an Olympus BX51 fluorescence microscope (Olympus America) equipped with a DP50 camera (numerical aperture of the objective lens, 1/120; Olympus Singapore), at ×400 magnification, and with Viewfinder Lite 1.0 image acquisition software (Better Light, Inc., San Carlos, CA). Preparations were scanned using spectral filters at wavelengths of 465, 510, 578, and 620 nm. Acquired spectral images were analyzed in a MATLAB software environment with programs written in MATLAB script language (Mathworks, Natick, MA).
Multispectral Image and Linear Mixture Model
The multispectral image of a specimen is represented by the LMM in a space induced by rth-order RVM (see Cover6), ϕ(X) (the mapping itself is formally defined in the next section):
| (1) |
where stands for the multispectral image comprised of I 3 spectral bands and I 1×I 2 pixels and stands for the matrix of sources representing spatial distributions of J materials present in the image. The first-order mapping represents the original image itself, and model (1) becomes a standard LMM in the original input space, X ≈ AS, used commonly in multispectral image analysis,7–11 where stands for a matrix of spectral profiles of J materials present in the image. In mapped space, and , where and . Hence, RVM has dimensionality (band) expansion effect. In the image segmentation problem considered, ϕ(X), A ϕ, and ϕ(S) are assumed to be non-negative. Matrix representation (1) is obtained from three-dimensional multispectral image tensor by row or column stacking procedures.
To obtain factorization (1) unique up to permutation and scaling indeterminacies, which are inherent to blind decompositions, sparseness constraints are imposed by non-negative matrix factorization (NMF) algorithms24 or statistical independence constraints are imposed by independent component analysis (ICA) algorithms25 on rows of ϕ(S). Sparseness implies that one material dominantly occupies each pixel footprint, and that condition is fulfilled for histopathological images of the specimen. The statistical independence assumption fails when materials are spectrally similar. This occurs, for example, in the case of low-dimensional multispectral imaging of a skin tumor with a low fluorescence intensity,10,11 but occurs also with multispectral microscopy imaging of an unstained specimen with low contrast between the cell and the background.
An alternative to matrix representation of multispectral image is tensor representation (see Ref. 13), with elements ; that is, the multispectral image is a set of I 3 spectral band images with the size of I 1×I 2 pixels. This is a standard notation adopted for use in multiway analysis.26 For the purpose of multispectral image decomposition, a Tucker3 tensor model is adopted27:
| (2) |
where, as in model (1), the image is represented in the mapped space and first-order mapping of ϕ yields two-dimensional LMM in the original space,13 thus: . Here, is a core tensor, are factors, and x n denotes n-mode product of a tensor with a matrix A (n). In mapped space, the dimension that corresponds to spectral mode I 3 is replaced by induced dimension , where . In tensor model (2), array factor corresponds to the matrix of spectral profiles A ϕ in model (1).13 In contrast to matrix factorization model (1), tensor factorization model (2) exploits spatial structure of the multispectral image and enables decomposition in which uniqueness does not depend on the fulfillment of hard constraints on model factors such as sparseness or statistical independence.18,24 Furthermore, spatial distributions of the materials present in the image (cell and background) are immediately obtained in tensor format13:
| (3) |
where and † denotes the Moore-Penrose pseudo-inverse.
Contrast Enhancement of Unstained Specimen by Rational Variety Mapping
With LMM-based representation of the multispectral image, it is apparent that contrast enhancement between the materials occurs when the angle between their spectral profiles (column vectors in the mixing matrix) is increased. This coincides with increased dimensionality of the space induced by rth-order RVM. The nonlinear band expansion, introduced in Refs. 10, 14, and 28, is just a special case (corresponding to r = 2) of the rth-order RVM. The rth-order RVM of the pixel vector pattern at spatial coordinate i 1 i 2 is of the form
| (4) |
That is, it involves monomials up to the order r. It increases dimensionality of the pattern from I 3 to . This, for I 3 = 4 spectral images used in the experiments reported here yields for RVM2 and for RVM3. This enables separability between mapped patterns with high probability, because the number of separating surfaces grows with r and is given with , where d = (I 3 + r)!I 3!r! [see equation (5) in Ref. 6]. However, computational complexity grows rapidly when r increases, and therefore second-order or perhaps third-order RVM are of primary practical interest. From the standpoint of pattern separability, order of the RVM depends on the strength of nonlinearities present in the image. Hence, it is probable that performance improvement brought by higher-order RVM will be small, compared with the increase in computational complexity. Nevertheless, RVM enables nonlinear blind multispectral image decomposition, wherein nonlinearities do not need to be known. Because of increased dimensionality of the mapped space, RVM also improves the contrast between spectrally similar materials (as for unstained specimen images, for which contrast is low). Nonetheless, it is important to note that factorization of the LMM of models (1) and/or (2) in mapped space does not immediately lead to the solution of the segmentation problem. That is because ϕ(S) in (1) resembles ϕ(X) in (4), implying that actual solutions are hidden among many spurious ones. However, mutual sparseness constraint simplifies ϕ(S), because all of the monomials that involve cross-products vanish and ϕ(S) becomes
| (5) |
where T denotes transpose. Now, sparseness-based factorization enforces factorization: where , represent equivalent sources that are mutually sparse (although different powers of the same source are not). For image segmentation this solution is legitimate, because any combination of the powers of the spatial distributions of the material is itself a spatial distribution of the material.
Factorization Methods for Unsupervised Segmentation of Multispectral Image
Here, we briefly review four state of-the-art methods for factorization of matrix (1) and tensor (2) models of the multispectral image of a principally unstained specimen. These methods are referred to as i) dependent component analysis (DCA),10,11 ii) multilayer hierarchical alternating least-square non-negative matrix factorization (HALS NMF),16,17 iii) non-negative matrix underapproximation (NMU),15 and iv) non-negative tensor factorization (NTF).18
DCA is an extension of independent component analysis for problems in which statistical independence assumption between sources is not fulfilled. Then, high-pass filtering based preprocessing transform T is applied on multispectral image (1), yielding T(ϕ(X)) = A ϕ T(ϕ(S)), whereupon rows of T(ϕ(S)) are more statistically independent than rows of ϕ(S). ICA algorithms are then applied to T(ϕ(X)), yielding more accurate estimation of A ϕ. Afterward, sources are recovered through . Candidates for preprocessing transform T include wavelet packets,29 filter banks,30 and innovations.31
The HALS NMF algorithm minimizes global cost function to estimate mixing matrix A ϕ, and a set of local cost functions , , to estimate the sources , where and underlining denotes the row vectors. To obtain factorization unique up to permutation and scaling, sparseness constraints are imposed on .17,12 As already noted, they enforce all of the powers of the same source to combine into one equivalent source: . An additional performance improvement of the NMF algorithms is obtained when they are applied in the multilayer mode.16
The NMU has been introduced recently as a refinement of the NMF algorithms toward sparse factorization of ϕ(X) in (1). In addition to non-negativity constraints imposed on A ϕ and ϕ(S), the cost function is minimized, imposing an underapproximation constraint on A ϕ and ϕ(S): A ϕϕ(S) ≤ ϕ(X). This naturally generates a sparse solution without imposing direct constraints on the rows of ϕ(S). Thus, problems associated with selection of optimal values of the regularization constants and/or number of layers are avoided. Depending on the norm of the cost function, NMU algorithm can be used in two versions: NMU-l2 if l2 norm is used, and NMU-l1 if l1 norm is used. Code for the NMU algorithm as used for image segmentation in the present study is available at http://www.core.ucl.ac.be/∼ngillis/papers/recursiveNMU.m.
The NTF algorithms are based on minimization of a chosen discrepancy measure between multispectral image tensor and its model (2). Discrepancy measures based on α- and β-divergences have been used recently, because of their adaptability to data statistics.24 Results related to unsupervised segmentation of the multispectral microscopy images of unstained specimens of nerve cells and spleen tissues reported here were obtained by an NTF algorithm that minimizes the β-divergence between and its model . For equations of the multiplicative NTF algorithms based on β-divergence, as well as α-divergence, we refer the reader to Ref. 18 and to sections 7.4.4 and 7.4.5 in Ref. 24.
Results
Contrast enhancement capability of the RVM-based methods was demonstrated on one multispectral fluorescent image of the unstained specimen of sciatic nerve fibers and one of the spleen tissue. Recorded multispectral images comprised four spectral images at 465, 510, 578, and 620 nm; 510 nm is the wavelength at which the best contrast was obtained; Figure 1A shows the 510-nm spectral image of unstained sciatic nerve fibers, and Figure 1B shows the cross-section of unstained spleen tissue. The cross-section of the spleen shows both red and white pulp; the white pulp consists further of lymph nodes or follicles. The image also shows a part of the spleen that is wrapped around the white pulp. This part consists of reticular fibers and is full of red blood cells. Part of the white pulp, which is of interest to our research, shows diffuse lymph tissue that belongs to the family of T lymphocytes.
Figure 1.
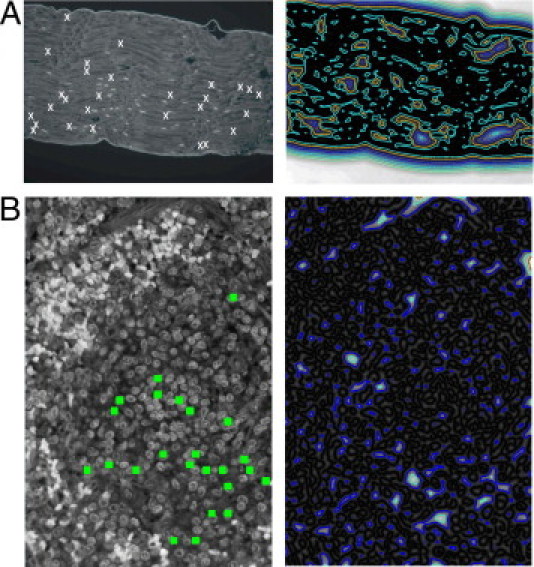
Spectral fluorescent microscopy images at 510 nm of histological slices of unstained specimens of sciatic nerve fibers (nervus ischiadicus) (A) and spleen (B). False-positive predicted nerve fiber spots (A) are marked by white crosses and false-positive predicted lymph nodes in the spleen tissue (B) are marked by green squares. Left: Spectral image. Right: Isolines of the estimated energy of the vector field convolution (VFC).
Two pathophysiologists (M.H. and M.P.H.) evaluated these images. In the image of unstained sciatic nerve fibers, both pathophysiologists detected 55 true-positive nerve fiber spots and 28 false-positive spots (Figure 1A). It is of greater importance, however, that an automated recognition would be highly unreliable because of weak contrast between the nerve spots and the background. This is demonstrated in Figure 1A, which shows isolines of the estimated external energy of the vector field convolution (VFC),32,33 calculated for the 510-nm spectral image of the unstained specimen. Because of weak boundaries, isolines are not closed around the spots but are scattered randomly across the image. Figure 2A shows images segmented from multispectral image of unstained specimen by the RVM2 transform and DCA algorithm and by the RVM3 transform and NMU-ℓ1 algorithm, with corresponding images obtained without RVM transform. Isolines of the estimated external energy (Figure 2B) are shown in one-to-one correspondence with their related images (Figure 2A).
Figure 2.
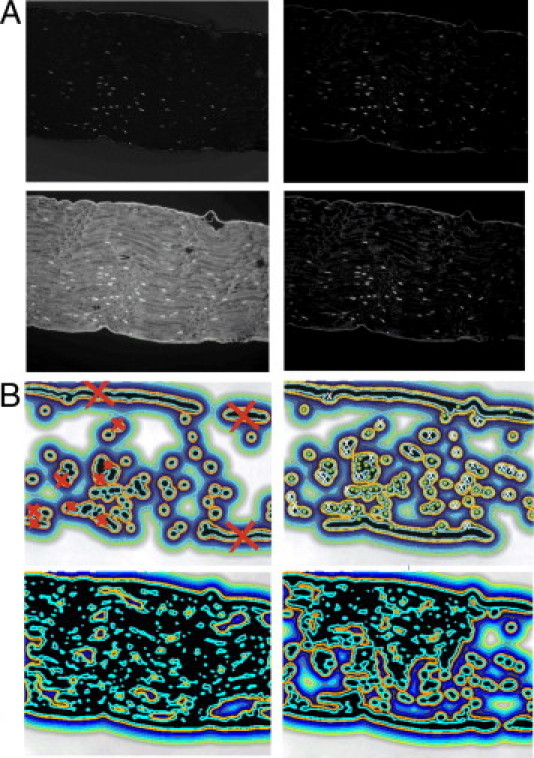
Segmented images of sciatic nerve fibers (A) with corresponding isolines of the estimated external energy of the VFC (B). Left: Second-order RVM2 and DCA algorithm. Right: Third-order RVM3 and NMU-ℓ1 algorithm. Isoline images in B are obtained from segmented images shown in A and are in one-to-one correspondence with them. On the isoline images obtained from the RVM-transformed images, the false-positive and false-negative spots are marked by red or white crosses, respectively; spots that fall on tissue border were excluded from the accuracy analysis.
The most important finding is that contrast enhancement due to the use of the RVM yielded stronger boundaries of the nerve fiber spots, and these are, therefore, better encircled by isolines. This enables more accurate classification and/or analysis of histopathological microscopy images. This statement is further supported by quantitative performance analysis performed by the two trained pathophysiologists (M.H. and M.P.H.). The numbers of true-positive (N TP), false-positive (N FP), and false-negative (N FN) outcomes are reported in Table 1 for the unstained image and for images segmented by the RVM2 and RVM3 transforms paired with DCA, β-NTF, HALS NMF, and NMU-ℓ1 algorithms. Table 2 reports sensitivity and positive predictive value. In the validation phase, the attention of the pathophysiologists was on the spots encircled by isolines, which would normally be checked by an automated classification system. Where a false positive or false negative fell in a part of the specimen corresponding to a border line (Figure 2B), it was excluded from the accuracy analysis (Table 2).
Table 1.
Outcomes for Image Analysis of the Unstained Specimen and of Images Segmented by the Different Transforms and Algorithms
| Outcome | Unstained specimen | RVM2 DCA | RVM3 DCA | RVM2 β-NTF | RVM3 β-NTF | RVM2 HALS NMF | RVM3 HALS NMF | RVM2 NMU-ℓ1 | RVM3 NMU-ℓ1 |
|---|---|---|---|---|---|---|---|---|---|
| Sciatic nerve fiber (nervus ischiadicus) | |||||||||
| NTP | 55/55 | 50/51 | 52/51 | 48/48 | 53/55 | 50/49 | 54/54 | 48/49 | 58/58 |
| NFN | 0/0 | 4/4 | 4/2 | 5/5 | 2/1 | 5/55 | 1/2 | 4/4 | 1/1 |
| NFP | 27/27 | 11/11 | 30/32 | 20/19 | 42/40 | 20/22 | 20/19 | 27/25 | 29/30 |
| Spleen tissue | |||||||||
| NTP | 128/131 | 128/131 | 126/121 | 128/131 | Failed | 128/131 | Failed | 128/131 | 118/117 |
| NFN | 0/0 | 3/7 | 3/5 | 3/6 | Failed | 5/5 | Failed | 5/8 | 2/1 |
| NFP | 55/47 | 41/48 | 24/23 | 24/33 | Failed | 31/25 | Failed | 29/35 | 17/19 |
Results obtained independently by two pathologists are separated by a virgule mark.
Transforms: RVM2 and RVM3 (second-order and third-order rational variety mapping). Algorithms: DCA (dependent component analysis), β-NTF (β-divergence non-negative tensor factorization), HALS NMF (hierarchical alternating least-square non-negative matrix factorization), and NMU-ℓ1 (non-negative matrix underapproximation).
NFN, number of false negatives; NFP, number of false positives; NTP, number of true positives.
Table 2.
Sensitivity and Positive Predictive Value of Image Analysis of the Unstained Specimen and of Images Segmented by the Different Transforms and Algorithms
| Statistic⁎ | Unstained specimen | RVM2 DCA | RVM3 DCA | RVM2 β-NTF | RVM3 β-NTF | RVM2 HALS NMF | RVM3 HALS NMF | RVM2 NMU-ℓ1 | RVM3 NMU-ℓ1 |
|---|---|---|---|---|---|---|---|---|---|
| Nerve fiber (nervus ischiadicus) | |||||||||
| Sens, % | 100/100 | 92.6/92.7 | 92.9/96.2 | 90.6/90.6 | 96.4/98.2 | 90.9/90.7 | 98.2/96.4 | 92.3/92.5 | 98.3/98.3 |
| PPV, % | 67/67 | 82/82.3 | 63.4/61.4 | 70.6/71.6 | 55.8/57.9 | 71.4/69 | 73/74 | 64/66.2 | 66.7/65.9 |
| Spleen tissue | |||||||||
| Sens, % | 97.7/97.7 | 97.7/94.9 | 97.7/96 | 97.7/95.6 | Failed | 96.2/96.3 | Failed | 96.2/94.2 | 98.3/99.2 |
| PPV, % | 70/73.6 | 75.7/73.2 | 84/84 | 84.2/79.9 | Failed | 80.5/84 | Failed | 81.5/78.9 | 87.4/86 |
Results obtained independently by two pathologists are separated by a virgule mark. Bold type indicates the several best results that combine high sensitivity and low false positive alarm rate.
Sens = NTP/(NTP + NFN) and PPV = NTP/(NTP + NFP), where Sens is sensitivity, N is number, TP is true positive, FN is false negative, FP is false positive, and PPV is positive predictive value.
The RVM2-DCA method yielded the smallest false-alarm rate (PPV ≈ 82.1%, with sensitivity (Sens) ≈ 92.6%; Table 2), which is significantly better than the rate obtained from the image of the unstained specimen. The highest accuracy (Sens ≈ 97.3%) was obtained with the RVM3-HALS NMF algorithm, but with increased false-alarm rate. Overall, the RVM2-DCA would be the method of choice for the present example. It is also important to note that, for the case of sciatic nerve fiber, the interobserver variability was small; the worst case was 3% for images segmented by RVM3-DCA and RVM2 NMU-ℓ1 algorithms. Figure 3 shows results obtained by the RVM2 and RVM3 transforms and NMU-ℓ1 algorithm, with corresponding isolines. Red blood cells located in the upper left corner are suppressed and the white pulp part of the spleen tissue is emphasized (Figure 3A). For a reference, an image of the unstained specimen and the corresponding isolines are shown in Figure 1B. Because of weak boundaries, isolines are scattered randomly across the image for the unstained specimen (Figure 1). Isolines shown in Figure 3B show more regular structure.
Figure 3.
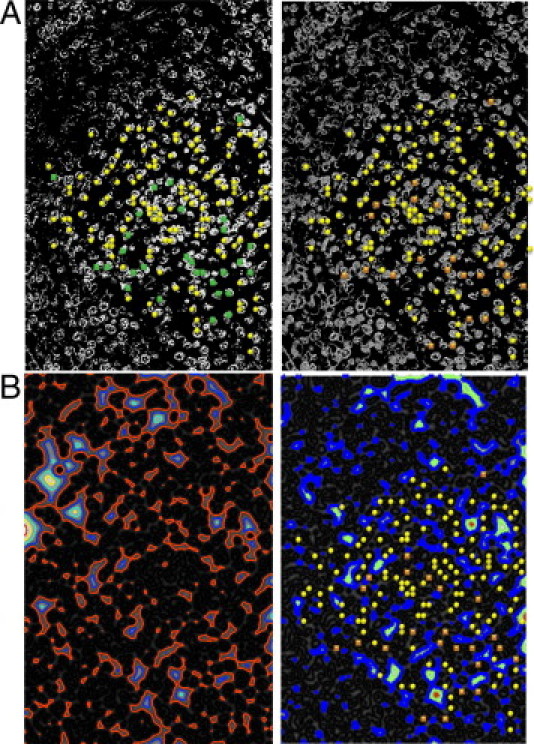
Segmented images of spleen tissue (A), with corresponding isolines of the estimated external energy of the VFC (B). Left: Second-order RVM2 NMU-ℓ1 algorithm. Right: Third-order RVM3 and NMU-ℓ1 algorithm. True-positive spots are marked with yellow circles. False-negative spots are marked with green squares in the second-order RVM2 NMU-ℓ1 segmented image and with orange squares in the third-order RVM3 NMU-ℓ1 segmented image.
Quantitative performance analysis was conducted by two pathophysiologists. The numbers of true-positive (N TP), false-positive (N FP), and false-negative (N FN) outcomes are reported in Table 1, and the sensitivity and positive predictive values are reported in Table 2. RVM3 paired with β-NTF and HALS NMF methods failed to detect true-positive spots in spleen tissue. These methods were therefore excluded from the accuracy analysis (Table 2). The failure was a consequence of significantly increased computational complexity of RVM3 and β-NTF/HALS NMF; it takes many hours to segment the image, and selection of the optimal values of free parameters becomes intractable. On the other hand, segmentation by RVM3 and DCA/NMU was performed within several minutes.
All methods yielded consistent improvement of false-alarm rate (≥10% improvement), relative to the case of image of unstained specimen (Table 2). Moreover, RVM3-NMU yielded the smallest false-alarm rate (PPV ≈ 86.7%) and the best accuracy (Sens ≈ 98.7%). In the present study, RVM3 brought additional performance improvement over RVM2, as did the RVM3-DCA algorithm.
Discussion
A contrast-enhancement methodology is proposed for nonlinear unsupervised segmentation of fluorescent multispectral microscopy images of unstained specimens. The methodology combines rth-order RVM, to increase spectral diversity between the materials present in the image, with non-negative matrix or tensor factorization. The methodology was demonstrated on images of unstained specimen of sciatic nerve fiber and spleen tissue. Lymphatic cells present in the spleen tissue were visually and anatomically clearly separated from their envelopes of red pulp. The results obtained imply that staining of the specimen only for contrast enhancement can be avoided, enabling the specimen to be used for visual inspection by a pathophysiologist, as well as for other purposes that require absence of staining. Moreover, the demonstrated contrast-enhancement enables design of an automated system for classification and analysis of unstained microscope histopathological images. RVM-based nonlinear blind image segmentation has two main advantages: it implicitly takes into account the possibly nonlinear nature of the image and it enhances contrast between spectrally similar materials, which occurs because of increased dimensionality of the mapped space. It is conjectured that a theoretical framework related to reproducible kernel Hilbert space will further be relevant for this purpose.
Footnotes
Supported by grants 098-0982903-2558 and 098-0982464-2460 funded by the Ministry of Science, Education and Sports, Republic of Croatia.
References
- 1.Lai C., Khosla R. GA based optimisation of a multi-agent soft computing model for segmentation and classification of unstained mammalian cell images. Proceedings of the 2003Congress of Evolutionary Computation 2003 (CEC 2003); 8–12 Dec. 2003, Canberra, Australia; Los Alamitos, CA: IEEE Computer Society Press; 2003. pp. 1192–1198. 2. [Google Scholar]
- 2.Lupica G., Allinson N.M., Botchway S.W. 2008: Hybrid image processing technique for robust identification of unstained cells in bright-field microscope images. Proceedings of the 2008International Conferences on Computational Intelligence for Modelling, Control and Automation; Intelligent Agents, Web Technologies and Internet Commerce; and Innovation in Software Engineering (CIMCA 2008, IAWTIC 2008, and ISE 2008); Vienna, Austria, Dec. 10–12, 2008; Los Alamitos, CA: IEEE Computer Society Press; 2008. pp. 1053–1058. 1. [Google Scholar]
- 3.Padfield D.R., Rittscher J., Sebastian T., Thomas N., Roysam B. Spatio-temporal cell cycle analysis using 3D level set segmentation of unstained nuclei in line scan confocal fluorescence images. Proceedings of the 2006 IEEE International Symposium on Biomedical Imaging: From Nano to Macro (ISBI 2006); Arlington, VA, April 6–9, 2006; Los Alamitos, CA: IEEE Computer Society Press; 2006. pp. 1036–1039. 1. [Google Scholar]
- 4.Tscherepanow M., Jensen N., Kummert F. Recognition of Unstained Live Drosophila Cells in Microscope Images. IEEE International Machine Vision and Image Processing Conference (IMVIP 2007); Maynooth, Ireland, Sept. 5–7, 2007; Los Alamitos, CA: IEEE Computer Society Press; 2007. pp. 169–176. 1. [Google Scholar]
- 5.Erasmus J.J., Gladish G.W., Broemeling L., Sabloff B.S., Truong M.T., Herbst R.S., Munden R.F. Interobserver and intraobserver variability in measurement of non-small-cell carcinoma lung lesions: implications for assessment of tumor response. J Clin Oncol. 2003;21:2574–2582. doi: 10.1200/JCO.2003.01.144. [DOI] [PubMed] [Google Scholar]
- 6.Cover T.M. Geometrical and statistical properties of systems of linear inequalities with applications in pattern recognition. IEEE Trans Elect Comput. 1965;EC-14:326–334. [Google Scholar]
- 7.Adams J.B., Smith M.O., Johnson P.E. Spectral mixture modeling: a new analysis of rock and soil types at the Viking Lander 1 suite. J Geophys Res. 1986;91:8098–8112. [Google Scholar]
- 8.Settle J.J., Drake N.A. Linear mixing and estimation of ground cover proportions. Int J Remote Sens. 1993;14:1159–1177. [Google Scholar]
- 9.Craig M. Minimum-volume transforms for remotely sensed data. IEEE Trans Geosci Remote Sens. 1994;32:542–552. [Google Scholar]
- 10.Kopriva I., Peršin A. Unsupervised decomposition of low-intensity low-dimensional multi-spectral fluorescent images for tumour demarcation. Med Image Anal. 2009;13:507–518. doi: 10.1016/j.media.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 11.Kopriva I., Peršin A., Puizina-Ivić N., Mirić L. Robust demarcation of basal cell carcinoma by dependent component analysis-based segmentation of multi-spectral fluorescence images. J Photochem Photobiol B. 2010;100:10–18. doi: 10.1016/j.jphotobiol.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 12.Kopriva I., Cichocki A. Blind decomposition of low-dimensional multi-spectral image by sparse component analysis. J Chemometrics. 2009;23:590–597. [Google Scholar]
- 13.Kopriva I., Cichocki A. Blind multispectral image decomposition by 3D nonnegative tensor factorization. Opt Lett. 2009;34:2210–2212. doi: 10.1364/OL.34.002210. [DOI] [PubMed] [Google Scholar]
- 14.Ouyang Y.C., Chen H.M., Chai J.W., Chen C.C.C., Poon S.K., Yang C.W., Lee S.K., Chang C.I. Band expansion-based over-complete independent component analysis for multispectral processing of magnetic resonance image. IEEE Trans Biomed Eng. 2008;55:1666–1677. doi: 10.1109/tbme.2008.919107. [DOI] [PubMed] [Google Scholar]
- 15.Gillis N., Glineur F. Using underapproximations for sparse nonnegative matrix factorization. Pattern Recog. 2010;43:1676–1687. [Google Scholar]
- 16.Cichocki A., Zdunek R. Multilayer nonnegative matrix factorization. Electron Lett. 2006;42:947–948. [Google Scholar]
- 17.Cichocki A., Zdunek R., Amari S.I. Hierarchical ALS algorithms for nonnegative matrix factorization and 3D tensor factorization. Independent Component Analysis; ICA07, London, UK, Sept. 9–12, 2007; Berlin: Springer; 2007. pp. 169–176. Lecture Notes Comput Sci 4666. [Google Scholar]
- 18.Cichocki A., Phan A.H. Fast local algorithms for large scale nonnegative matrix and tensor factorizations. IEICE Trans Fundamentals. 2009;E92:A708–A721. [Google Scholar]
- 19.Zimmer C., Labruyère E., Meas-Yedid V., Guillén N., Olivio-Marin J.C. Segmentation and tracking of migrating cells in videomicroscopy with parametric active contours: a tool for cell-based drug testing. IEEE Trans Med Imaging. 2002;21:1212–1221. doi: 10.1109/TMI.2002.806292. [DOI] [PubMed] [Google Scholar]
- 20.Arambula Cosio F., Márquez Flores J.A., Padilla Castañeda M.A., Solano S., Tato P. Automatic counting of immunocytochemically stained cells: Engineering in Medicine and Biology Society, 2003. Proceedings of the 25th Annual International Conference of the IEEE; Cancun, Mexico, Sept. 17–21, 2003; Los Alamitos, CA: IEEE Computer Society Press; 2003. pp. 790–793. 1. [Google Scholar]
- 21.Masmoudi H., Hewitt S.M., Petrick N., Myers K.J., Gavrielides M.A. Automated quantitative assessment of HER-2/neu immunohistochemical expression in breast cancer. IEEE Trans Med Imaging. 2009;28:916–925. doi: 10.1109/TMI.2009.2012901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mao K.Z., Zhao P., Tan P.H. Supervised learning-based cell image segmentation for p53 immunohistochemistry. IEEE Trans Biomed Eng. 2006;53:1153–1163. doi: 10.1109/TBME.2006.873538. [DOI] [PubMed] [Google Scholar]
- 23.Arif M., Rajpoot N. Classification of potential nuclei in prostate histology images using shape manifold learning. International Conference on Machine Vision (ICMV 2007); Islamabad, Pakistan, Dec. 28–29, 2007; Los Alamitos, CA: IEEE Computer Society Press; 2007. pp. 113–118. 1. [Google Scholar]
- 24.Cichocki A., Zdunek R., Phan A.H., Amari S.I. John Wiley; Chichester, UK: 2009. Alternating Least Squares and Related Algorithms for NMF and SCA Problems: Nonnegative Matrix and Tensor Factorizations: Applications to Exploratory Multi-Way Data Analysis and Blind Source Separation, ch 4; pp. 203–266. [Google Scholar]
- 25.Cichocki A., Amari S.I. John Wiley; Chichester, UK: 2002. Adaptive Blind Signal and Image Processing; pp. 129–174. 231–233. [Google Scholar]
- 26.Kiers H.A.L. Towards a standardized notation and terminology in multiway analysis. J Chemometrics. 2000;14:105–122. [Google Scholar]
- 27.Tucker R.L. Some mathematical notes on three-mode factor analysis. Psychometrika. 1966;31:279–311. doi: 10.1007/BF02289464. [DOI] [PubMed] [Google Scholar]
- 28.Du Q., Kopriva I., Szu H. Independent component analysis for classifying multispectral images with dimensionality limitation. Int J Inf Acquis. 2004;1:201–216. [Google Scholar]
- 29.Kopriva I., Seršić D. Wavelet packets approach to blind separation of statistically dependent sources. Neurocomputing. 2008;71:1642–1655. [Google Scholar]
- 30.Tanaka T., Cichocki A. Subband decomposition independent component analysis and new performance criteria. Proceedings of the IEEE Conference on Acoustics, Speech, and Signal Processing; 2004(ICASSP '04); Los Alamitos, CA: IEEE Computer Society Press; 2004. pp. 541–544. 5. [Google Scholar]
- 31.Hyvärinen A. Independent component analysis for time-dependent stochastic processes. Proceedings of the International Conference on Artificial Neural Networks (ICANN '98) 1998; Skovde, Sweden, Sept. 2–4, 1998; Berlin: Springer; 1998. pp. 541–546. 1. [Google Scholar]
- 32.Li B., Acton S.T. Active contour external force using vector field convolution for image segmentation. IEEE Trans Image Process. 2007;16:2096–2106. doi: 10.1109/tip.2007.899601. [DOI] [PubMed] [Google Scholar]
- 33.Li B., Acton S.T. Automatic active model initialization via Poisson inverse gradient. IEEE Trans Image Process. 2008;17:1406–1420. doi: 10.1109/TIP.2008.925375. [DOI] [PubMed] [Google Scholar]


