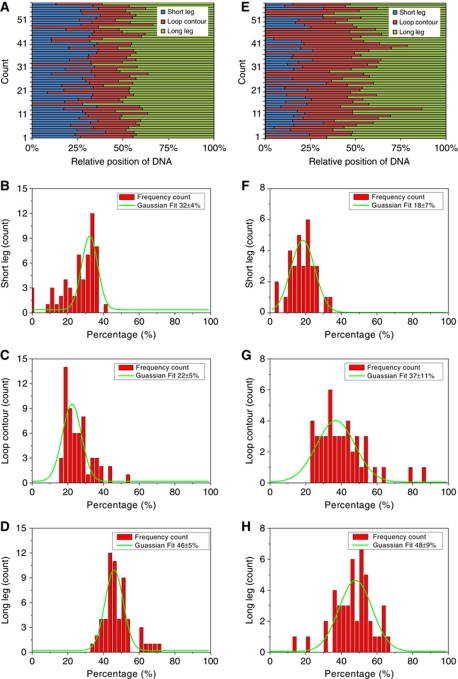
Figure 4.
Normalized length distributions and Gaussian fits for MutS bound in loop configuration to heteroduplex DNA containing biotin at both DNA ends. MutS (100 nM) and 1120-bp G-T mismatch DNA (20 nM) that contained biotin at both ends and that had been previously bound with monovalent streptavidin (see Materials and methods) were incubated in 20 mM Tris–HCl (pH 7.5), 100 mM K+ glutamate, 5 mM MgCl2, and 0.4 mM DTT for 10 min at room temperature in the absence (A–D) or presence (E–H) of 0.5 mM ATP. Panels (A) and (E) show the contour lengths of each segment on individual DNA molecules. In each diagram, the left (blue) bar indicates the contour length of the shortest segment of DNA between MutS and the end of the DNA, the middle bar (red) indicates the contour length of the loop, and the right bar (green) indicates the contour length of the DNA segment between MutS and the other DNA end. The length of each segment was normalized to the total DNA length. The histograms below (B–D) and (F–H) show Gaussian fits of the contour length data. In the presence of ATP, the peak of the Gaussian distribution of the lengths of the short legs (the distance from the base of the loop to the nearest DNA end) decreased from 32 to 17% of the DNA length and the peak of the Gaussian distribution of the loop length increased from 22 to 36% of the DNA length, while the peak of the Gaussian distribution of the lengths of the long legs (the distance from the base of the loop to the furthest DNA end) remained at 46–47% of the DNA length.