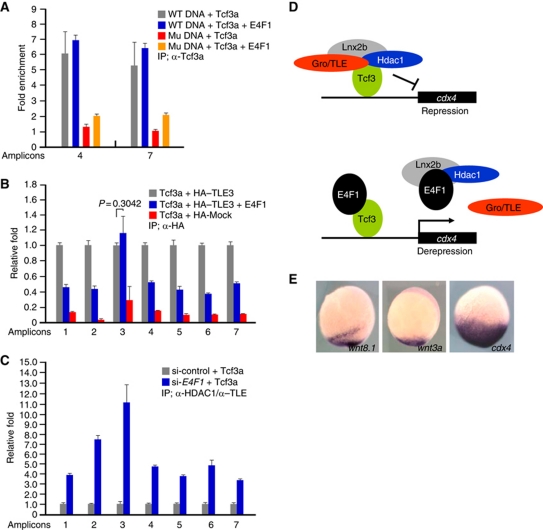
Figure 7.
Tcf3 remains bound to the cdx4 promoter in the presence of E4f1 while the corepressors are removed. (A–C) ChIP analyses in 293T cells transfected with the indicated DNAs. Amplicons are identified in Figure 4A. (A) PCR amplified DNA fragments including WT or mutant cdx4 regulatory elements (Supplementary Figure S9Aa and d, respectively) were co-transfected with Tcf3a with or without E4F1, as indicated. Samples were immunoprecipitated with anti-Tcf3a antibody, or IgG as control. Amplicons 4 and 7 were used for q-PCR. Fold enrichment is calculated setting the IgG control as one. Note that E4F1 did not interfere with Tcf3a binding to the DNA, while mutant DNA was recovered at low yield. (B) ChIP q-PCR assay showing that E4F1 dissociates TLE3 from the Tcf3-binding sites in the cdx4 promoter; the aberrant behaviour of amplicon 3 is not understood. (C) ChIP q-PCR analysis of cdx4 regulatory region after IP with anti-HDAC1 and anti-TLE antibodies after depleting endogenously expressed E4F1. cdx4-luc plasmid and Tcf3a were co-transfected with or without E4F1 siRNA. Note that ChIP yields increased significantly in E4F1-siRNA-treated cells. q-PCR was carried out at least in triplicate, and standard error of the mean is shown in the panels. A proposed mechanism is shown in (D). In this model, a complex composed of Tcf3–Gro/TLE–HDAC1–Lnx2b represses the cdx4 gene; E4f1 dissociates the complex, preserving Tcf3 binding to the DNA. (E) Expression of wnt3a and wnt8.1 is much more restricted in the caudal domain at the bud stage than cdx4. A proposed role for E4f1 derepression in achieving this pattern is discussed in the text.