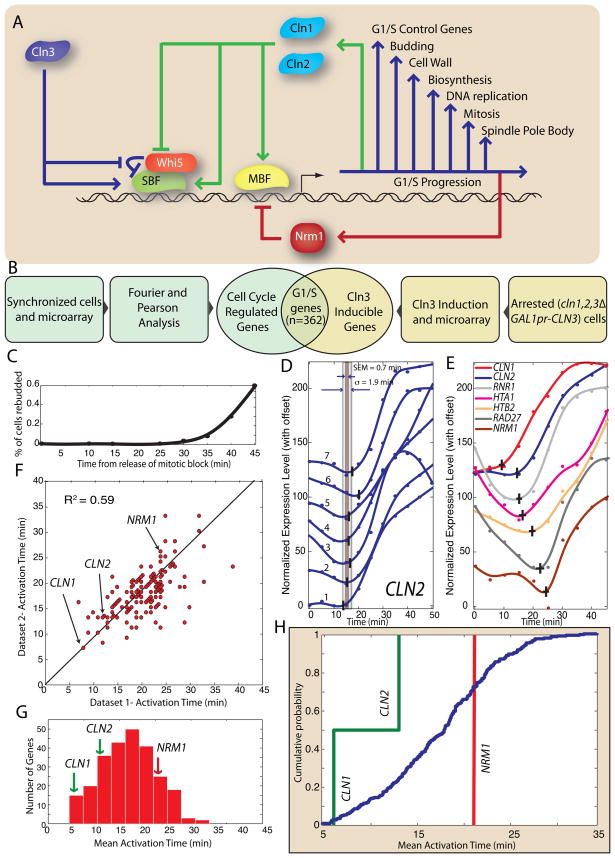
Figure 1. Positive feedback precedes genome-wide change in transcription at G1/S in S. cerevisiae.
(A) Schematic diagram of the G1/S transition. (B) The G1/S regulon is defined as the intersection of the set of cell cycle regulated genes with the set of Cln3-inducible genes. (C) Synchrony of cdc20Δ GALLpr-CDC20 metaphase block-release from Di Talia et al (2009). (D) An algorithm is applied to a smoothing-spline fit to detect activation of CLN2 transcription in 7 mitotic block-release datasets (See Figures S1-3 for algorithm description; specific genotypes of Datasets 1–7 described in Figure S1D). The standard deviation σ and the standard error of the mean (SEM) is calculated for each gene. (E) 7 genes in the G1/S regulon are activated at different times; data shown from a single dataset. The vertical and horizontal bars indicate the activation time and its SEM respectively. (F) Gene activation time correlation between two of the 7 datasets (R2 = 0.59; see supplementary information for additional correlations). Histogram (G) and corresponding cumulative distribution (H) of mean activation times for the 7 mitotic block-release datasets. CLN1 and CLN2, two genes responsible for positive feedback, are among the earliest-activated genes. NRM1, a negative regulator of MBF, is activated later.