Abstract
Centromeres nucleate the formation of kinetochores and are vital for chromosome segregation during mitosis. The SNF2 family helicase PICH (Plk1-interacting checkpoint helicase) and the BLM (the Bloom's syndrome protein) helicase decorate ultrafine histone-negative DNA threads that link the segregating sister centromeres during anaphase. The functions of PICH and BLM at these threads are not understood, however. Here, we show that PICH binds to BLM and enables BLM localization to anaphase centromeric threads. PICH- or BLM-RNAi cells fail to resolve these threads in anaphase. The fragmented threads form centromeric-chromatin-containing micronuclei in daughter cells. Anaphase threads in PICH- and BLM-RNAi cells contain histones and centromere markers. Recombinant purified PICH has nucleosome remodelling activities in vitro. We propose that PICH and BLM unravel centromeric chromatin and keep anaphase DNA threads mostly free of nucleosomes, thus allowing these threads to span long distances between rapidly segregating centromeres without breakage and providing a spatiotemporal window for their resolution.
Keywords: centromere, chromatin remodelling, DNA repair, mitosis
Introduction
Centromeres are special regions of chromosomes that are crucial for chromosome segregation in mitosis. On one hand, centromeric chromatin nucleates kinetochore formation in mitosis (Cleveland et al, 2003; Przewloka and Glover, 2009). Kinetochores then serve as the spindle microtubule attachment sites and are signalling hubs for the spindle checkpoint, a cellular surveillance system that ensures the fidelity of chromosome segregation (Cleveland et al, 2003; Bharadwaj and Yu, 2004; Musacchio and Salmon, 2007; Yu, 2007). On the other hand, because of the balance between cohesin removal and protection mechanisms in prophase, centromeres are the major sites of cohesin binding at metaphase (Watanabe, 2005). Centromeric cohesion resists the spindle pulling force exerted at the kinetochores and prevents premature sister-chromatid separation. Therefore, centromeres orchestrate opposing mitotic processes and coordinate chromosome segregation in mitosis.
Mammalian centromeres contain up to megabases of tandem satellite DNA repeats (Buscaino et al, 2010; Dalal and Bui, 2010). Centromeric DNA is neither necessary nor sufficient for the formation of functional centromeres. Epigenetic mechanisms contribute significantly to centromere identity. These mechanisms include characteristic histone modifications and the incorporation of the histone H3-like protein CENP-A (in place of histone H3) into a fraction of nucleosomes in centromeric chromatin (Buscaino et al, 2010; Dalal and Bui, 2010).
Plk1-interacting checkpoint helicase (PICH) was discovered as a binding protein of the mitotic kinase Plk1 (Baumann et al, 2007; Leng et al, 2008). It contains an N-terminal DNA helicase domain (HD) that is related to the SNF2 family of helicases, which is generally involved in chromatin remodelling and does not catalyse DNA strand separation (Saha et al, 2006; Hopfner and Michaelis, 2007). Plk1 binds to PICH through a phosphorylated Ser-Ser-Pro motif at the C-terminal region of PICH (Baumann et al, 2007; Leng et al, 2008). PICH localizes to kinetochores during prometaphase and decorates ultrafine DNA threads in anaphase. When cells are directly fixed without manipulations, these threads are devoid of histones, cannot be stained with DNA intercalating dyes such as DAPI (4′,6-diamidino-2-phenylindole), and frequently connect separated centromere pairs (Baumann et al, 2007; Wang et al, 2008). Because inactivation of topoisomerase IIα (Topo IIα) increases the frequency and persistence of PICH-positive threads in cells, it is highly likely that PICH can decorate catenated centromeric DNA (Spence et al, 2007; Wang et al, 2008). PICH was also proposed to function in the spindle checkpoint (hence its name) and to regulate the kinetochore localization of Mad2, a key spindle checkpoint protein (Baumann et al, 2007). The spindle checkpoint function of PICH was recently called into question, however (Hubner et al, 2010). Finally, PICH has been shown to regulate abscission (the last step of cytokinesis) and chromosome architecture (Kurasawa and Yu-Lee, 2010).
The Bloom's syndrome protein (BLM) is a DNA helicase that unwinds double-stranded DNA and other types of DNA structures (Chu and Hickson, 2009). BLM is mutated in Bloom's syndrome patients, who develop cancers at a young age, among other phenotypes. BLM has multiple critical roles in DNA repair (Chu and Hickson, 2009). First, BLM suppresses homologous recombination (HR) by disrupting the formation of displacement loops, a key step in HR (Bachrati et al, 2006; Bugreev et al, 2007). Second, as a component of protein complex that also consists of topoisomerase IIIα (Topo IIIα), RMI1/BLAP75, and RMI2, BLM promotes the dissolution of double Holliday junctions to produce non-crossover HR products (Yin et al, 2005; Raynard et al, 2006, 2008; Wu et al, 2006; Singh et al, 2008; Xu et al, 2008). As a result, a hallmark of BLM-deficient cells is the elevated frequency of sister-chromatid exchanges (SCEs) (Chaganti et al, 1974; Luo et al, 2000), which are crossover products of HR. The second hallmark of BLM-deficient cells is the formation of micronuclei (Rosin and German, 1985). It is unclear whether and how the functions of BLM in suppressing SCE and in preventing micronuclei formation are linked. Interestingly, BLM and its associated proteins Topo IIIα and RMI1 also decorate centromeric anaphase DNA threads that are positive for PICH (Chan et al, 2007). The function of BLM at these DNA threads is unknown.
In this study, we identify BLM as a binding partner of PICH. Unlike BLM depletion, PICH depletion does not elevate SCE, suggesting that PICH might not be required for the DNA repair function of BLM in interphase. PICH- and BLM-RNAi cells fail to resolve anaphase threads properly, leading to their breakage and micronuclei formation in daughter cells. Consistent with the fact that the majority of PICH- and BLM-positive anaphase threads connect segregating centromeres, about 50% of the micronuclei in PICH- and BLM-RNAi cells contain centromere markers. Most importantly, the unresolved anaphase threads in PICH-RNAi and BLM-RNAi cells contain histones. Furthermore, recombinant PICH has nucleosome remodelling activity in vitro. We propose that PICH and BLM collaborate to unravel centromeric chromatin into fully extended and stretched DNA threads and maintain these threads in a nucleosome-free state, thus allowing them to span long distances between rapidly segregating sister centromeres during anaphase without breakage. This mechanism then affords a spatiotemporal window for Topo II and other DNA repair enzymes to access and resolve catenated or other aberrant DNA structures at the midpoint of these threads.
Results
Identification of BLM as a PICH-binding protein
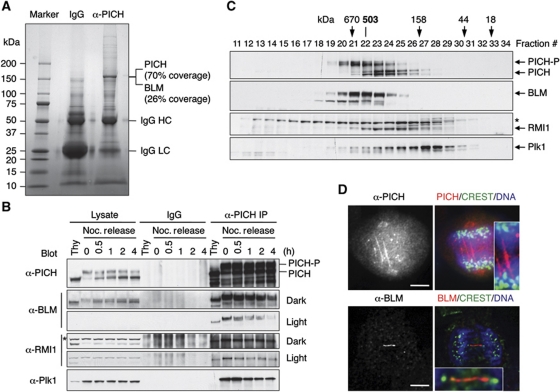
We first confirmed that multiple siRNAs against PICH depleted Mad2 through off-target effects in HeLa cells, and that PICH was not required for the spindle checkpoint (data not shown). To explore the functions of PICH, we immunoprecipitated the endogenous PICH from log-phase HeLa cells using an anti-PICH antibody (Figure 1A). Mass spectrometry analysis revealed that the major band present in the anti-PICH immunoprecipitates (IP) contained PICH and BLM. The sequence coverage of PICH and BLM by mass spectrometry was 70 and 26%, respectively. Immunoblotting confirmed that BLM was indeed present in anti-PICH IP (Figure 1B). The PICH–BLM interaction was present in G1/S, increased in mitosis, and decreased during mitotic exit (Figure 1B). PICH also interacted with Plk1 and became hyperphosphorylated in mitosis, confirming published reports (Baumann et al, 2007; Leng et al, 2008).
Figure 1.
Identification of BLM as a PICH-binding protein. (A) Control IgG and anti-PICH IP from HeLa Tet-On cells were resolved by SDS–PAGE and stained with colloidal Coomassie blue. The molecular mass of markers is labelled. The sequence coverage of PICH and BLM in mass spectrometry is indicated. (B) Lysates, control IgG IP, or anti-PICH IP of HeLa Tet-On cells arrested in G1/S by thymidine (Thy) or at different times (hours) following the release from nocodazole-triggered mitotic arrest were blotted with the indicated antibodies. A cross-reacting band in the α-RMI1 blot is indicated with an asterisk. The positions of hyperphosphorylated and hypophosphorylated PICH are labelled. Light exposures of the α-BLM and α-RMI1 blots are included to better reveal their decrease in PICH binding during mitotic exit. (C) Lysates of nocodazole-arrested mitotic HeLa cells were fractionated on a Superose 6 column. The fractions were analysed by SDS–PAGE followed by immunoblotting with the indicated antibodies. The positions of hypophosphorylated and hyperphosphorylated forms of PICH are labelled. The faster migrating band in the α-BLM blot is derived from BLM, as it was depleted by BLM siRNA. A cross-reacting band in the α-RMI1 blot is indicated with an asterisk. The elution positions of the molecular mass standards are indicated. (D) HeLa Tet-On cells in anaphase were stained with anti-PICH or anti-BLM along with CREST (a centromere marker) and DAPI (stains DNA). In the overlay images, anti-PICH/BLM staining is in red. CREST staining is in green. DAPI staining is in blue. The PICH and BLM threads are magnified in the insets. Scale bars, 5 μm.
BLM forms a complex with Topo IIIα, RMI1, and RMI2, some of which also decorate anaphase threads (Chan et al, 2007). RMI1 was present in anti-PICH IP in all cell cycle stages tested (Figure 1B). The endogenous PICH, BLM, and RMI1 in HeLa cells co-fractionated on a gel filtration column (Figure 1C). The hyperphosphorylated PICH appeared to co-fractionate more precisely with BLM. The native molecular mass of this complex was 503 kDa, which fitted well with a calculated molecular mass of 498 kDa of a complex containing one molecule each of PICH, BLM, Topo IIIα, RMI1, and RMI2. Therefore, although our mass spectrometry analysis failed to identify Topo IIIα, RMI1, or RMI2 in anti-PICH IP from log-phase cells, RMI1 and possibly other BLM-binding proteins interacted with PICH in mitosis. There were no good commercially available antibodies against Topo IIIα and RMI2, precluding us from testing their binding to PICH. The bulk of Plk1 did not co-fractionate with the PICH–BLM complex (Figure 1C). A recent proteomic study also identified components of the BLM complex as PICH-interacting proteins in mitosis (Hutchins et al, 2010).
To test whether PICH bound to BLM in the absence of other components of the complex, we co-expressed PICH and BLM in Sf9 insect cells. Recombinant PICH and BLM co-fractionated on a gel filtration column at a native molecular mass of 340 kDa, consistent with the two forming a 1:1 heterodimer (Supplementary Figure S1). Furthermore, recombinant PICH and BLM could be co-immunoprecipitated. Therefore, PICH and BLM directly interacted with each other in this heterologous system.
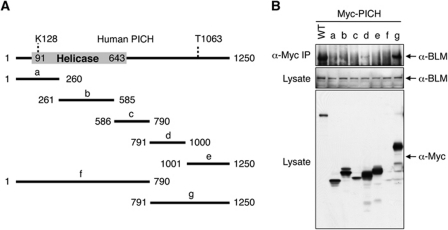
PICH contains an N-terminal SNF2 HD and a C-terminal non-HD. To further map the BLM-binding region of PICH, we constructed several Myc-PICH truncation mutants, expressed them in HeLa cells, and tested their interactions with the endogenous BLM (Figure 2). As expected, the full-length Myc-PICH interacted with endogenous BLM. The C-terminal non-HD of PICH, Myc-PICHg, also interacted with BLM, suggesting that the C-terminal non-HD mediates BLM binding. Even though it was expressed at much higher levels, Myc-PICHg pulled down less BLM than did full-length Myc-PICH, suggesting that the N-terminal HD of PICH also contributed to BLM binding. We could not ascertain whether the N-terminal HD of PICH on its own bound to BLM, because the fragment encompassing the HD, Myc-PICHf, did not express well in HeLa cells. Regardless, these results further confirmed a physical interaction between PICH and BLM.
Figure 2.
The C-terminal region of PICH binds to BLM. (A) Schematic drawing of the domain structure of human PICH. K128 is critical for ATP binding and for the helicase activity of PICH. Phosphorylation of T1063 creates a docking site for Plk1. The boundaries of the PICH truncation mutants are shown. (B) HeLa Tet-On cells were transfected with plasmids encoding either wild-type (WT) Myc-PICH or Myc-PICH truncation mutants described in (A). Lysates and anti-Myc IP of these cells were blotted with the indicated antibodies.
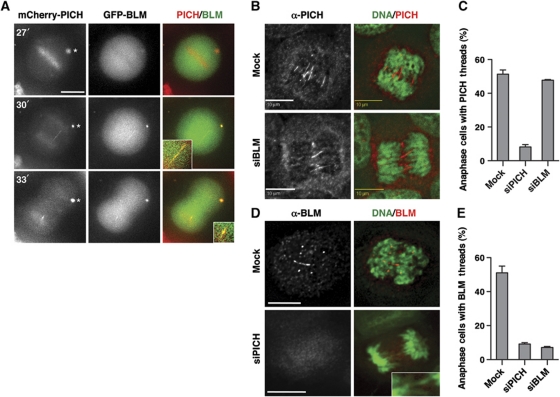
PICH and BLM co-localize to ultrafine DNA threads in anaphase (Chan et al, 2007). We confirmed that PICH localized to kinetochores during prometaphase and to fine threads in anaphase (Figure 1D; Supplementary Figure S2). We also observed BLM localization to anaphase threads (Figure 1D). These PICH and BLM threads were DAPI-negative and frequently connected centromeres (Figure 1D). PICH and BLM also formed foci on anaphase chromosomes that did not stain with the centromere marker CREST (Figure 1D; see also Figure 5B and D, below). The nature of these foci was not further investigated, but they might be related to the fragile site foci that were positive for Fanconi anaemia proteins (Chan et al, 2009; Naim and Rosselli, 2009).
PICH depletion does not increase SCE
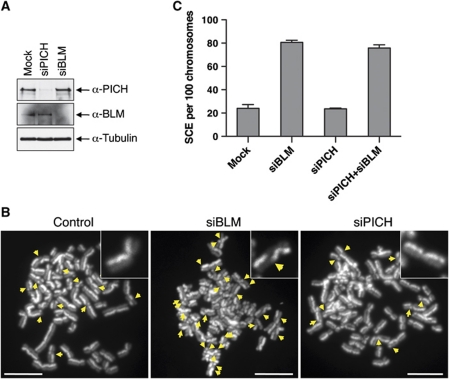
BLM-deficient cells exhibit two major phenotypes: (1) elevated SCE and (2) micronuclei formation (Chu and Hickson, 2009). We first tested whether PICH was required for suppressing SCE. As expected, depletion of BLM in HeLa cells markedly elevated the frequency of SCE (Figure 3). By contrast, depletion of PICH did not alter the SCE frequency. In this and all subsequent PICH-RNAi experiment, we primarily used siPICH4, as it did not deplete Mad2. Co-depletion of PICH and BLM did not further increase SCE frequency as compared with BLM depletion alone. Consistently, PICH and BLM did not co-localize well in interphase cells (Supplementary Figure S3). Therefore, PICH does not appear to be required for the function of BLM in suppressing SCE, although we cannot rule out the possibility that the residual amount of PICH in PICH-RNAi cells is sufficient to perform this function.
Figure 3.
PICH is not required for suppressing SCE. (A) Lysates of HeLa Tet-On cells mock transfected or transfected with siPICH or siBLM were blotted with the indicated antibodies. (B) Representative metaphase spreads from the SCE assays of mock RNAi, BLM-RNAi, and PICH-RNAi HeLa Tet-On cells. The SCE events are indicated with arrows. A sister-chromatid pair from each metaphase spread is magnified and shown in insets. Scale bars, 10 μm. (C) Quantification of the number of SCE events per 100 chromosomes in cells either mock transfected or transfected with siBLM, siPICH, or both. Mean and s.d. of three independent experiments are shown.
PICH depletion increases the frequency of micronuclei formation
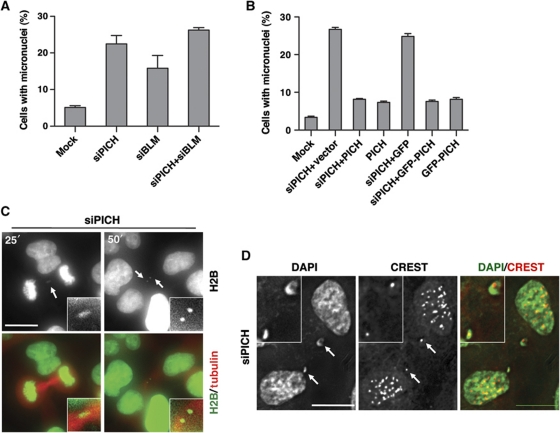
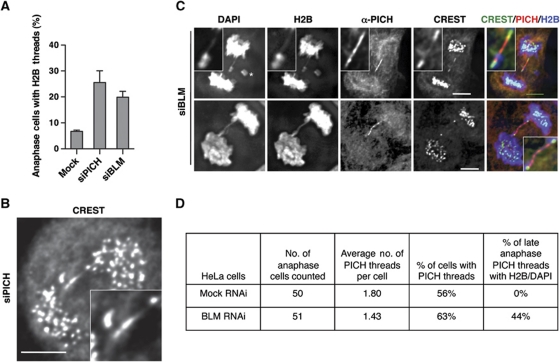
We next examined whether PICH was required to prevent micronuclei formation. Depletion of BLM expectedly caused micronuclei formation in about 20% of the cells (Figure 4A; Supplementary Figure S4A). Depletion of PICH also resulted in micronuclei formation in about 20–30% of the cells. Furthermore, co-depletion of PICH and BLM did not markedly increase the percentage of cells containing micronuclei (Figure 4A), suggesting that PICH and BLM might function in the same pathway to suppress micronuclei formation. Importantly, ectopic expression of RNAi-resistant untagged or GFP-tagged PICH proteins greatly reduced the percentage of micronuclei-positive PICH-RNAi cells (Figure 4B; Supplementary Figure S4B). We also created stable HeLa Tet-On cell lines that expressed siRNA-resistant Myc-PICH driven by the doxycycline-inducible promoter. Induced expression of Myc-PICH at a level lower than that of the endogenous PICH also partially rescued the micronuclei phenotype caused by siPICH4 transfection (Supplementary Figure S4C and D). These results indicate that the observed micronuclei phenotype of PICH-RNAi cells requires inactivation of the PICH protein and is not due to off-target effects. Moreover, depletion of Topo IIIα, RMI1, or RMI2 by RNAi also caused micronuclei formation (Supplementary Figure S4E). Therefore, PICH is required for suppressing micronuclei formation and possibly acts in the same pathway as do BLM and other components in the complex.
Figure 4.
PICH prevents formation of centromere-positive micronuclei. (A) Quantification of the percentage of micronuclei-containing HeLa Tet-On cells either mock transfected or transfected with siPICH, siBLM, or both. Mean and s.d. of three independent experiments are shown. (B) Ectopic expression of untagged PICH or GFP-PICH rescues the micronuclei phenotype of PICH-RNAi cells. Quantification of the percentage of micronuclei-containing HeLa Tet-On cells mock transfected or transfected with siPICH together with empty vector or plasmids encoding GFP or siPICH4-resistant PICH or GFP-PICH. Mean and s.d. of three independent experiments are shown. (C) Live cell imaging of PICH-RNAi HeLa Tet-On cells expressing H2B-mCherry (pseudo-coloured green) and GFP-tubulin (coloured red). Images at 25 and 60 min after NEBD are shown. Images from additional time points are shown in Supplementary Figure S6B. The H2B thread and micronuclei are magnified and shown in insets. Scale bar, 10 μm. (D) Representative images of PICH-RNAi cells stained with DAPI (green in overlay) and CREST (red in overlay). Two CREST-positive micronuclei are magnified and shown in insets. Scale bars, 10 μm.
To further probe how micronuclei form in PICH- and BLM-RNAi cells, we performed live cell imaging experiments. HeLa cells stably expressing H2B-mCherry and GFP-tubulin were transfected with siRNAs against PICH or BLM and monitored with time-lapse microscopy. Mock-transfected cells performed mitosis normally, rarely forming micronuclei (Supplementary Figure S5A). PICH-RNAi cells, however, often contained thin H2B-mCherry threads at the cleavage furrow in anaphase (Supplementary Figure S5B; Figure 4C). As illustrated in Figure 4C, a representative PICH-RNAi cell contained a short H2B-mCherry thread in anaphase at 25 min after mitotic entry. This thread broke, producing two micronuclei in telophase (at 50 min after mitotic entry). Each daughter inherited one micronucleus. Similar fashions of micronuclei formation were observed for BLM-RNAi cells (Supplementary Figure S5C). Thus, one mechanism of micronuclei formation in PICH- and BLM-RNAi cells is the breakage of thin chromatin threads. Consistently, two adjacent cells that both contained micronuclei were frequently observed in immunostaining of fixed cells (Supplementary Figure S4A). These two cells were likely the two daughter cells resulting from the same cell division.
Because both PICH and BLM localized to DNA threads in anaphase, it was possible that their inactivation led to a failure to resolve these threads. Consistent with the fact that the majority of the PICH- and BLM-positive threads were derived from centromeres (Baumann et al, 2007; Chan et al, 2007), about 50% of the micronuclei in control, PICH-, and BLM-RNAi cells were positive for the centromere marker, CREST (Figure 4D). Therefore, our results suggest that PICH and BLM localize to centromeric DNA threads in anaphase and collaborate to resolve these threads.
PICH is required for BLM recruitment to anaphase threads
We examined the kinetics of PICH and BLM recruitment to anaphase threads using live cell imaging. HeLa cells were transfected with mCherry-PICH and GFP-BLM and observed with time-lapse microscopy (Figure 5A). Similar to the endogenous PICH, mCherry-PICH localized to chromosomes at prometaphase and metaphase while GFP-BLM was largely absent from chromosomes at these stages. Thus, PICH localization to chromosomes preceded that of BLM. At 3 min after anaphase onset, both mCherry-PICH and GFP-BLM co-localized to an anaphase thread. The PICH/BLM-positive thread disappeared 3 min later and became a single PICH/BLM focus. This result suggests that the localization of PICH and BLM to anaphase threads and such threads are transient.
Figure 5.
PICH is required for BLM localization to anaphase threads. (A) Live cell imaging of HeLa Tet-On cells transfected with plasmids encoding mCherry-PICH (red in overlay) and GFP-BLM (green). Images at 27, 30, and 33 min after NEBD are shown. The PICH/BLM-positive thread and foci are magnified and shown in insets. Note that overexpressed PICH tended to form cytoplasmic foci (indicated with an asterisk), which also contained BLM at the 30- and 33-min time points. Scale bar, 10 μm. (B) Mock-transfected or BLM-RNAi HeLa Tet-On cells were stained with anti-PICH (red in overlay) and DAPI (green in overlay). Scale bars, 10 μm. (C) Quantification of the percentage of anaphase cells transfected with the indicated siRNAs that contained PICH threads. Mean and s.d. of three independent experiments are shown. (D) Mock-transfected or PICH-RNAi HeLa Tet-On cells were stained with anti-BLM (red in overlay) and DAPI (green in overlay). A DAPI-positive anaphase thread is shown in inset. Scale bars, 10 μm. (E) Quantification of the percentage of anaphase cells transfected with the indicated siRNAs that contained BLM threads. Mean and s.d. of three independent experiments are shown.
We next tested whether PICH and BLM depend on each other for localization to anaphase threads. Depletion of BLM did not affect the percentage of anaphase cells that contained PICH-positive threads (Figure 5B and C). Thus, BLM is dispensable for PICH localization to anaphase threads, consistent with previous reports (Chan et al, 2007). By contrast, depletion of PICH markedly decreased the percentage of anaphase cells that had BLM-positive threads (Figure 5D and E). The representative PICH-RNAi cell contained DAPI-positive threads, but did not have BLM-positive threads (Figure 5D). Therefore, PICH is required for BLM localization to anaphase threads.
Anaphase threads in PICH- and BLM-RNAi cells contain histones
In unperturbed anaphase cells and without further manipulation, the PICH/BLM-positive threads lack histones and cannot be stained with DAPI. Surprisingly, we were able to observe micronuclei formation in PICH- and BLM-RNAi cells by monitoring H2B-mCherry (Figure 4C; Supplementary Figure S5). This finding suggested that the unresolved threads in PICH- and BLM-RNAi cells contained histones. To confirm this finding, we fixed PICH-RNAi H2B-GFP cells and stained them with DAPI, CREST, and anti-PICH. About 25% of PICH-RNAi cells in anaphase contained threads that were positive for histones (Figure 6A). In rare cases, we observed anaphase threads decorated by centromere markers not only at the ends but also along its length (Figure 6B), suggesting that the unresolved anaphase threads in PICH-RNAi cells contained stretched centromeric chromatin. This type of centromeric threads was not observed in control cells. We also stained BLM-RNAi H2B-GFP cells with DAPI, CREST, and anti-PICH. Similar to PICH-RNAi cells, about 20% of BLM-RNAi anaphase cells contained histone-positive threads (Figure 6A). The percentages of anaphase PICH- and BLM-RNAi cells containing H2B threads correlated well with the percentages of these cells that formed micronuclei (see Figure 4A).
Figure 6.
PICH and BLM prevent histone association with anaphase threads. (A) Quantification of the percentage of H2B-thread-containing anaphase HeLa Tet-On cells stably expressing H2B-GFP transfected with the indicated siRNAs. (B) A representative anaphase PICH-RNAi cell stained with CREST. An anaphase thread decorated with CREST is magnified and shown in inset. Scale bar, 5 μm. (C) Two representative anaphase BLM-RNAi H2B-GFP-expressing cells stained with DAPI, anti-PICH, and CREST. CREST, PICH, and H2B signals in the overlay are coloured green, red, and blue, respectively. PICH threads are magnified and shown in insets. Scale bars, 5 μm. (D) Statistics of the number of PICH threads and the percentage of PICH threads with histone H2B and DAPI in mock or BLM-RNAi anaphase HeLa cells.
Because BLM was not required for PICH localization to anaphase threads, we were able to visualize the PICH threads in BLM-RNAi cells. About 44% (37 out of 84) PICH-positive threads contained histones whereas none of the PICH threads in control cells contained histones or DAPI (Figure 6C and D; Supplementary Figure S6A), indicating that BLM was required to prevent histone association with these threads. Because the two major segregating masses of chromosomes overlapped during early anaphase, it was difficult to ascertain whether PICH threads contained histones at this stage or they simply overlapped with the bulk of histone signals by chance. For this reason, we only analysed PICH threads in late anaphase to determine if they contained histones. Importantly, all PICH threads with histones could also be stained with DAPI (Figure 6C).
Certain PICH threads in BLM-RNAi cells were stained with centromere markers in their entire length (Figure 6C, top panel). Finally, PICH-positive threads in BLM-RNAi cells persisted till telophase, even after chromosome decondensation (Supplementary Figure S6B). These threads were non-contiguous and exhibited punctuated staining of centromere markers along their length. Our findings are consistent with a role of the PICH–BLM complex in remodelling centromeric chromatin and in preventing the association of histones and possibly CENP-A with these threads. In the absence of their functions, the anaphase threads are packaged into centromeric chromatin and cannot be efficiently resolved before breakage. Finally, other mechanisms likely contribute to micronuclei formation. For example, the BLM-RNAi cell contained a lagging chromosome fragment without centromeres (Figure 6C, top panel; indicated with an asterisk), which may form a micronucleus without centromere markers in the daughter cell.
We next examined the PICH threads in BLM-deficient cells (PSNG13) and in stable transfectants of BLM-deficient cells that expressed a functional BLM transgene (PSNF5). Compared with PSNG13 cells, PSNF5 cells had fewer SCE events per chromosome (Figure 7A), confirming that PSNF5 cells had functional BLM. Consistent with a previous study, PSNF5 cells contained fewer PICH threads per anaphase cells (Figure 7B). By contrast, there was no appreciable difference in the number of PICH threads between control and BLM-RNAi HeLa cells (Figure 6D). The underlying reason for this discrepancy was not clear. One possibility was that BLM depletion in HeLa cells was incomplete. The residual BLM was sufficient to promote the resolution of some PICH-positive threads, which failed to accumulate in BLM-RNAi cells.
Figure 7.
BLM is required for anaphase thread resolution. (A) Quantification of SCE in PSNG13 (BLM−) and PSNF5 (BLM+) cells. (B) Statistics of the number of all PICH threads and the number of DAPI-positive PICH threads in anaphase PSNG13 (BLM−) and PSNF5 (BLM+) cells. (C) Representative images of anaphase PSNG13 (BLM−) and PSNF5 (BLM+) cells. The PICH thread in the PSNG13 cell contained DAPI while the PICH thread in the PSNF5 cell did not. Scale bars, 5 μm.
Importantly, the BLM-deficient cells (PSNG13) contained more and higher percentage of PICH threads that were positive for DAPI (Figure 7B and C), as compared with PSNF5 cells, confirming the results in BLM-RNAi HeLa cells. We could not co-stain these cells with both antibodies against PICH and histones, as the commercially available monoclonal antibodies against bulk histones did not work in IF. On the other hand, all PICH-/DAPI-positive threads in BLM-RNAi cells contained H2B-GFP, and vice versa. Thus, the PICH-/DAPI-positive threads in BLM-deficient PSNG13 cells likely contained histones.
Recombinant PICH has nucleosome remodelling activity
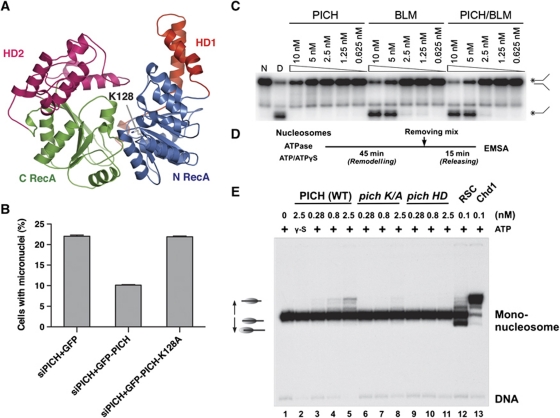
PICH has an SNF2 HD. Structural modelling with Rad54 as the template suggested that the HD of PICH adopts a classical SNF2 helicase fold with two RecA-like lobes (N RecA and C RecA) and two helical domains (HD1 and HD2) (Thoma et al, 2005) (Figure 8A). Because K128 of PICH in the glycine-rich loop is critical for ATP binding, mutations of K128 are expected to diminish the helicase activity of PICH. Ectopic expression of GFP-PICH wild type (WT), but not GFP-PICH K128A, effectively rescued the micronuclei formation phenotype of PICH-RNAi cells (Figure 8B). This result indicates that the helicase activity of PICH is required to prevent micronuclei formation. Consistently, this helicase-deficient PICH K128A mutant was less efficient in forming anaphase threads when the endogenous PICH was depleted (Supplementary Figure S7).
Figure 8.
PICH has nucleosome sliding activity. (A) Ribbon diagram of a homology-based structure model of the HD of PICH. The model is built with the structure of Rad54 (PDB ID: 1Z3I) as the template. The ATP-binding residue K128 is shown in sticks. (B) Quantification of the percentage of micronuclei-containing HeLa Tet-On cells transfected with siPICH4 together with plasmids encoding GFP, siPICH4-resistant GFP-PICH, or GFP-PICH K128A. Mean and s.d. of three independent experiments are shown. (C) Helicase assay with PICH, BLM, and the PICH–BLM complex. Native (N) replication-fork-like duplex DNA and denatured (D) single-stranded DNA were included as controls. (D) Reaction schemes of the nucleosome sliding assay. (E) Nucleosome sliding assay of wild-type (WT) PICH, PICH K128A (K/A), PICH HD, RSC, and Chd1.
The SNF2 family of helicases is typically involved in chromatin remodelling by translocating on DNA and does not generally catalyse strand separation of duplex DNA. We first verified that PICH indeed did not catalyse strand separation of duplex DNA in a classical helicase assay (Figure 8C). PICH also did not stimulate the helicase activity of BLM in this assay. We thus tested whether PICH had nucleosome remodelling activity. We incubated PICH WT, PICH K128A, and the HD of PICH (PICH HD) with reconstituted nucleosomes that contained a 32P-labelled DNA fragment in the presence of ATP or ATPγS (a non-hydrolyzable ATP analogue), and subjected the reaction mixtures to electrophoretic mobility shift assay (Figure 8D and E). As positive controls in this assay, the RSC (remodelling structure of chromatin) complex and Chd1 (chromodomain helicase DNA-binding protein 1) efficiently remodelled nucleosomes. RSC primarily moved nucleosomes to border positions on the DNA (Figure 8E, lane 12) whereas Chd1 (an SNF2 family helicase) repositioned nucleosomes towards the centre of the DNA (Figure 8E, lane 13). PICH WT exhibited dose-dependent nucleosome remodelling activity in this assay that required ATP hydrolysis (Figure 8E, lanes 2–5). As expected, this activity was greatly diminished in the PICH K128A mutant (Figure 8E, lanes 6–8). The HD of PICH (PICH HD) alone was also inactive (Figure 8E, lanes 9–11), indicating that the C-terminal non-HD of PICH was required for this activity. Furthermore, similar to Chd1, PICH moved nucleosomes towards central positions on DNA. Taken together, these results demonstrate that PICH is a chromatin remodelling enzyme.
Histone incorporation into anaphase threads also occurs in BLM-RNAi cells, indicating that BLM is required for the chromatin remodelling function of PICH in vivo. Furthermore, BLM is known to stimulate the nucleosome remodelling activity of Rad54, another SNF2 helicase, and this stimulation does not involve the helicase activity of BLM (Srivastava et al, 2009). We thus tested whether BLM stimulated the nucleosome remodelling activity of PICH. We co-expressed PICH and BLM (or its helicase-dead mutant; BLM HD) in Sf9 cells and partially purified the resulting complexes (Supplementary Figure S8A). The PICH–BLM complexes exhibited nucleosome remodelling activities similar to that of PICH alone (Supplementary Figure S8B). Thus, BLM binding does not stimulate the activity of PICH in vitro.
Discussion
Centromeres are vital for chromosome inheritance and stability. In this study, we have demonstrated a role of the PICH–BLM helicase duo in the maintenance of centromere integrity. Our results suggest that PICH and BLM collaborate to limit histone incorporation into fine DNA threads connecting the separating sister centromeres in anaphase, allowing these threads to stretch and span long distances without breakage (Figure 9). In so doing, they promote the resolution of these threads by providing a spatiotemporal window for DNA repair enzymes. Inactivation of this pathway leads to packaging of centromeric DNA in the ultrafine threads into chromatin fibres, resulting in the breakage of these DNA threads in telophase and micronuclei formation in daughter cells.
Figure 9.
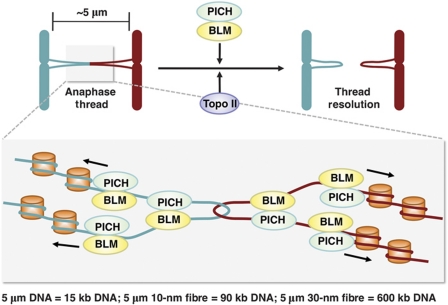
Model of PICH/BLM-dependent resolution of anaphase centromeric DNA threads. In this model, PICH and BLM collaborate to keep anaphase DNA threads largely free of nucleosomes, providing a spatiotemporal window for Topo II and possibly other DNA repair enzymes to resolve catenated DNA or other aberrant DNA structures.
Structure of the anaphase centromeric DNA threads
Three types of DNA structures have been proposed for these DNA threads: (1) concatenated DNA, (2) replication intermediates, and (3) recombination intermediates (e.g. Holliday junctions) (Chan et al, 2007; Wang et al, 2008). Induction of replication stress by aphidicolin caused a modest increase of the number of centromeric anaphase threads (Naim and Rosselli, 2009), suggesting a minor contribution of replication intermediates in the formation of these threads. RNAi depletion of Rad51, a key enzyme in HR, does not reduce the number of centromeric anaphase threads (YK and HY, unpublished results), which disfavours the involvement of recombination intermediates in these threads. Finally, inactivation of Topo II, an enzyme that catalyses DNA decatenation, greatly increases the number of centromeric PICH/BLM threads (Spence et al, 2007; Wang et al, 2008). Therefore, the available evidence suggests that most PICH/BLM-positive centromeric anaphase threads represent catenated DNA (Figure 9).
DNA catenation is formed as a result of DNA replication and must be resolved to allow proper sister-chromatid separation (Wang, 2002). DNA decatenation and cohesin removal need to be coordinated (Yanagida, 2009). Depletion of the centromeric cohesin protector Sgo1 by RNAi causes precocious dissociation of cohesin from centromeres and premature sister-chromatid separation (Kitajima et al, 2004; Salic et al, 2004; Tang et al, 2004, 2006; McGuinness et al, 2005). Sgo1-RNAi cells contain much more PICH-positive threads as compared with control cells (Baumann et al, 2007), suggesting that DNA decatenation cannot keep pace when cohesin removal is ahead of schedule. Conversely, cohesin removal might be required for complete decatenation of centromeric DNA (Wang et al, 2010). Therefore, it is possible that DNA decatenation has not been completed at certain centromeres when the sister centromeres have separated in anaphase. PICH and BLM then decorate these catenated DNA threads and promote their resolution in anaphase.
Some PICH/BLM-positive threads are derived from chromosome arms (Chan et al, 2009; Naim and Rosselli, 2009). These non-centromeric threads are also positive for FANC-D2. Formation of these threads is greatly enhanced by replication stress, suggesting that they might represent replication intermediates (Chan et al, 2009; Naim and Rosselli, 2009). BLM has been shown to have a role in the resolution of these threads following replication stress (Chan et al, 2009; Naim and Rosselli, 2009). Future studies will determine whether PICH is also required for this process.
Resolution of anaphase threads by PICH and BLM
In this study, we show that PICH and BLM are components of a large multisubunit complex in mitosis. BLM has multiple roles in DNA repair (Chu and Hickson, 2009). BLM-deficient cells exhibit several characteristic phenotypes, including elevated frequency of SCE and micronuclei formation. We show that, unlike BLM-RNAi cells, PICH-RNAi cells do not exhibit a higher frequency of SCE, suggesting that PICH might not regulate BLM function in interphase. Instead, PICH and BLM co-localize to and promote resolution of DNA threads connecting segregated sister centromeres in anaphase and are required for their resolution. Without proper functions of PICH and BLM, breakage of the unresolved threads in telophase leads to the formation of micronuclei in daughter cells. These micronuclei frequently contain centromere markers, consistent with the fact that >50% of the PICH/BLM-positive anaphase threads connect centromeres. Our studies suggest that PICH specifically regulates the mitotic function of BLM. Because PICH inactivation does not increase SCE, but yet causes micronuclei formation, the functions of BLM in suppressing SCE and in preventing micronuclei formation can be uncoupled.
The PICH–BLM complex as a chromatin remodelling enzyme
How do PICH and BLM promote the resolution of centromeric anaphase threads? In one model, PICH recruits the BLM–Topo IIIα–RMI1–RMI2 complex to anaphase threads. The BLM complex then resolves these catenated DNA threads. We disfavour this model for two reasons. First, although BLM and its associated Topo IIIα have been shown to unwind and dissolve many DNA structures, including double Holliday junctions and replication intermediates, there are no demonstrated functions for the BLM complex in DNA decatenation (Chu and Hickson, 2009). In fact, the BLM complex is unlikely to have decatenation activity against fully catenated DNA, as Topo IIIα is a type IA topoisomerase that only catalyses the passing of one DNA single strand through another (Wang, 2002). Second, DNA catenation or other aberrant DNA structures are expected to occur only at or near the midpoint of each thread (Figure 9). If the sole function of BLM is to directly act on these aberrant structures, it is not apparent why PICH and BLM need to decorate the entire length of the thread.
Centromeric DNA threads in unperturbed anaphase cells do not contain histones. Unexpectedly, PICH-/BLM-RNAi cells contain thin histone/CREST-positive chromatin threads that connect centromeres. In particular, when BLM function is compromised, PICH-positive threads contain histones and centromere markers. Our results suggest that PICH and BLM are required to unravel centromeric chromatin to produce or maintain the nucleosome-free state of centromeric DNA threads. PICH has nucleosome remodelling activity in vitro, and the helicase activity of PICH is required to prevent micronulei formation in vivo. Therefore, our results are more consistent with an alternative ‘chromatin unravelling’ model (Figure 9).
In this model, PICH, BLM, and other components in the complex collaborate to unravel centromeric chromatin and produce nucleosome-free, catenated DNA threads connecting segregating centromeres. They decorate these threads along their entire length to prevent histone association and nucleosome formation. The relaxation of centromeric chromatin allows mere 15 kb of DNA from each centromere to form a 5-μm loop and to readily span the distance of rapidly separating sister centromeres. This mechanism thus provides a temporal and spatial window for Topo II and other DNA repair enzymes to resolve DNA catenation and other aberrant DNA structures. Inactivation of the PICH–BLM pathway leads to failure to produce or maintain the nucleosome-free state of these DNA threads, causing their compaction into centromeric chromatin and increasing the possibility of their breakage before resolution.
This model is consistent with the fact that PICH and BLM are found on several types of anaphase threads that presumably contain different, aberrant DNA structures. This model also provides possible explanations for the necessity of PICH and BLM to decorate the entire length of the ultrafine DNA threads and for the roles of BLM and Topo IIIα in resolving catenated DNA, through their putative functions in stimulating the chromatin remodelling activity of PICH. BLM alone is insufficient to stimulate the activity of PICH in vitro, suggesting that other components of this complex or post-translational modifications or both might be required for this process. Our model is not mutually exclusive with the possibility that BLM as a part of this complex directly resolves certain aberrant DNA structures in anaphase threads, such as incompletely replicated DNA or recombination intermediates.
In conclusion, we have demonstrated a functional requirement for PICH and BLM in the resolution of centromeric anaphase threads and consequently in the maintenance of centromere integrity. We provide evidence to further suggest that PICH and BLM promote the resolution of DNA threads in anaphase by unravelling centromeric chromatin and limiting histone association. Our studies thus reveal hitherto underappreciated functions of DNA repair enzymes in remodelling condensed chromatin and in preventing DNA breakage in mitosis.
Materials and methods
Cell culture, transfection, and siRNAs
HeLa Tet-On (Invitrogen) cells were grown in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum and 10 mM L-glutamine. For cell-cycle arrest in G1/S or mitosis, cells were treated for 24 h with 2 mM thymidine or for 16 h with 100 ng/ml nocodazole, respectively. At 40–50% confluency, cells were transfected with plasmids or siRNAs with Effectene (Qiagen) or Lipofectamine RNAiMax (Invitrogen), respectively, according to the manufacturer's instructions. To establish stable cell lines, HeLa Tet-On cells were transfected with pTRE2-Myc-based plasmids encoding RNAi-resistant Myc-PICH4M (contain silent mutations resistant to siPICH4). Selection of stable clones was performed in the presence of 300 μg/ml hygromycin. The surviving clones were screened for the induced expression of Myc-PICH in the absence or presence of 1 μg/ml doxycycline (Invitrogen).
For RNAi experiments, the siRNAs were chemically synthesized at Dharmacon. The siRNA oligonucleotides used in this study are siPICH4, AGGCCAGACUUAAUGAAAAdTdT; siBLM, GGAGCACAUCUGUAAAUUAdTdT; siTopo IIIα, GAAACUAUCUGGAUGUGUAdTdT; siRMI1, GGCCCAAAUGAAUAAACAAdTdT; and siRMI2, GUAGAAGAUUUACACAGGAdTdT. The siRNAs were transfected at a final concentration of 1 nM.
Antibodies, immunoblotting, and immunoprecipitation
To generate antibodies against PICH, the fragment of PICH (residues 791–1000) was produced in bacteria as a His6-tagged fusion protein and purified. The protein was used to immunize rabbits at Yenzym Antibodies (South San Francisco, CA). The antisera were purified with Affi-Gel beads (Bio-Rad) coupled to the antigen. The production of anti-APC2 and anti-Mad2 antibodies was described previously (Fang et al, 1998; Tang et al, 2001). The commercial antibodies used in this study were as follows: CREST (ImmunoVision), anti-BLAP75/RMI1 (Abcam, ab70525), rabbit anti-BLM (Abcam, ab2179), goat anti-BLM (Santa Cruz, sc-7790), anti-Myc (Roche, 11667203001), anti-Plk1 (Santa Cruz, SC-17783). For immunoblotting and immunofluorescence, the antibodies were used at a final concentration of 1 μg/ml.
For immunoblotting, cells were lysed in SDS sample buffer, sonicated, boiled, separated by SDS–PAGE, and blotted with the indicated antibodies. Horseradish peroxidase-conjugated goat anti-rabbit or goat anti-mouse IgG (Amersham Biosciences) was used as the secondary antibodies, and immunoblots were developed using the ECL reagent (Amersham Biosciences) according to the manufacturer's protocols and exposed to film.
For immunoprecipitation, whole-cell lysate was prepared by lysing cells in NP-40 lysis buffer (50 mM Tris–HCl pH 7.7, 150 mM NaCl, 0.5% v/v Nonidet P-40, 1 mM DTT and 1 × protease inhibitor cocktail) on ice for 15 min, sonicating three times, then centrifuging the lysed cells at 16 000 g for 15 min at 4 °C. Affinity-purified anti-PICH or anti-Myc coupled to Affi-Prep Protein A beads (Bio-Rad) at a concentration of 1 mg/ml were incubated with the supernatants for 2 h at 4 °C. The beads were then washed five times with the NP-40 lysis buffer. The proteins bound to the beads were dissolved in SDS sample buffer, separated by SDS–PAGE, and blotted with the appropriate antibodies.
Immunofluorescence and live cell imaging
For immunofluorescence, HeLa Tet-On cells or HeLa Tet-On cells expressing H2B-GFP were plated in four-well chamber slides (LabTek) and treated as indicated. Cells were fixed with 4% paraformaldehyde in 250 mM HEPES pH 7.4, 0.1% Triton X-100 at 4 °C for 20 min. After 3–5 washes over 20 min in PBS, cells were permeabilized in 0.5% Triton X-100 for 20 min and then washed with PBS. The cells were blocked in PBS plus 5% fetal bovine serum followed by a 16-h incubation with the primary antibodies. After 3–5 PBS washes over 20 min, cells were incubated with fluorescent secondary antibodies (Alexa Fluor 488 or 647, Molecular Probes) for 30 min at room temperature. After incubation, cells were washed with PBS and their nuclei were stained with DAPI (1 μg/ml). Slides were mounted and viewed with a × 100 objective on a Deltavision microscope. All images were taken at 0.2 μm intervals, deconvolved, and stacked. The images were further processed in ImageJ and pseudo-coloured in Adobe Photoshop. For quantification, multiple random fields were captured and 50–200 cells were counted in each of the three independent experiments.
For live cell imaging, HeLa Tet-On cells stably expressing GFP-tubulin and H2B-mCherry were plated in four-well chamber coverslips (LabTek), transfected with indicated siRNAs against PICH or BLM for 48 h, synchronized at G1/S by thymidine arrest for 18 h and released into fresh medium for 5 h before filming. For visualizing BLM or PICH localization, HeLa Tet-On cells were transfected with plasmids encoding GFP-BLM and mCherry-PICH for 48 h, synchronized at G1/S by thymidine arrest for 18 h and released into fresh medium for 5 h before filming. Cells were imaged in CO2-independent medium (Invitrogen) at 37 °C in a humidified chamber using a Deltavision microscope. Three Z-stacks were acquired every 3 or 5 min for 24 h. Image manipulations (contrast enhancement, cropping, and conversion to QuickTime movies) were performed with ImageJ.
Nucleosome sliding assay
The RSC complex (Rsc2-TAP) and the Chd1-TAP were purified through the tandem-affinity purification approach from budding yeasts (Li et al, 2005). His6-tagged PICH, PICH K128A, PICH HD (residues 60–680), PICH–BLM, and PICH–BLM helicase-dead mutant were expressed in Sf9 insect cells and purified with the Ni2+-NTA beads (Qiagen). Mononucleosomes were reconstituted with a 32P-labelled 216 bp DNA including the 601 sequence through the octamer transfer method (Owen-Hughes et al, 1999), and gel purified as described previously (Li et al, 2007). Sliding assays were performed at 37 °C in the sliding buffer (20 mM HEPES pH 7.9, 50 mM KCl, 10 mM MgCl2, 0.5 mM PMSF, 2 mM DTT, 0.05% Igepal CA-830, 10% glycerol, and 100 μg/ml BSA) in the presence of 4 mM ATP or ATPγS. A removing mix of 750 ng calf thymus DNA and 500 ng oligonucleosomes was added to stop the reactions (Li et al, 2005). The reaction mixtures were resolved by a native polyacrylamide gel followed by autoradiography.
SCE assay
HeLa Tet-On cells were transfected with the indicated siRNAs for 24 h. The cells were replated and incubated in the presence of 100 μM BrdU for 30 h (two cell divisions). Colcemid (150 ng/ml) was added during the final 30 min to enrich for mitotic cells. Cells were collected by trypsinization and washed in PBS. Cells were swelled in 75 mM KCl for 16 min at 37 °C, followed by centrifugation. Cell pellets were resuspended in fixative (3:1 solution of methanol:glacial acetic acid) and incubated for 20 min at 4 °C. Cells were washed in fixative two more times. After the final wash, cells were resuspended in fixative and dropped onto cold slides. Slides were allowed to air dry in the dark for 2–3 days. Chromosomes were then differentially stained for 5 min with 0.1 mg/ml acridine orange (Molecular Probes). Slides were washed extensively for 2 min under running water followed by a 1-min incubation and mounting in Sorenson Buffer, pH 6.8 (0.1 M Na2HPO4, 0.1 M NaH2PO4). Slides were immediately viewed with a × 63 objective on a Zeiss Axiovert 200 M fluorescence microscope. Images were acquired with a CCD camera using Slidebook imaging software (Intelligent Imaging Innovations). Images were analysed for the number of SCE by counting the number of crossover events per chromosome. About 200 chromosomes were scored for each experiment. Three independent experiments were performed.
Helicase assay
Two 50 nt DNA oligonucleotides were designed to form a 25-bp partial duplex when annealed. Before annealing, one oligonucleotide was 5′ 32P-labelled with T4 polynucleotide kinase (Promega) and purified using a Sephadex G50 spin column (Ambion). In all, 10 pmol labelled, purified oligonucleotide was combined with 100 pmol complimentary oligonucleotide in an annealing buffer containing 10 mM Tris–HCl pH 7.4 and 10 mM MgCl2. The duplex substrate was diluted to 10 nM in annealing buffer and stored at −20 °C. PICH, BLM, or the PICH–BLM complex was serially diluted, and added to a helicase buffer containing 20 mM Tris–HCl pH 7.4, 2 mM MgCl2 2 mM ATP, 0.1 mg/ml UltraPure BSA (Ambion), 1 mM DTT, and 1 nM 32P-labelled replication-fork substrate. After a 30-min incubation at 37 °C, TBE DNA loading buffer was added. The samples were separated on a native 15% acrylamide TBE gel (Bio-Rad). The gel was dried and analysed with a phosphoimager.
Supplementary Material
Acknowledgments
We thank Weidong Wang for BLM-deficient cells, Laura Diaz-Martinez for suggesting the chromatin unravelling model, and other members of the Yu laboratory for helpful discussions. This work was supported by the National Institutes of Health (GM61542 and GM76481) and the Welch Foundation (I-1441). BL (in the Department of Molecular Biology) is a WA ‘Tex’ Moncrief, Jr Scholar in Medical Research, and is supported by the National Institute of Health (GM090077), the March of Dimes Foundation, the Welch Foundation (I-1713), and American Heart Association. HY is an Investigator with the Howard Hughes Medical Institute.
Author contributions: YK conceived, performed, and analysed experiments described in Figures 1–6, 8B, and Supplementary Figures S1–S6 and S8A. J-WH and BL in the Department of Molecular Biology performed and analysed experiments in Figure 8E and Supplementary Figure S8B. RW performed and analysed data included in Figures 7B, C, 8C and Supplementary Figure S7. BL in the Department of Pharmacology constructed PICH WT and K128A baculoviruses and purified recombinant PICH. NW performed experiments in Figure 7A. ML constructed PICH and K128A plasmids. JZ and HLB analysed the PICH–BLM complex with mass spectrometry. HY supervised all experiments and wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Bachrati CZ, Borts RH, Hickson ID (2006) Mobile D-loops are a preferred substrate for the Bloom's syndrome helicase. Nucleic Acids Res 34: 2269–2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann C, Korner R, Hofmann K, Nigg EA (2007) PICH, a centromere-associated SNF2 family ATPase, is regulated by Plk1 and required for the spindle checkpoint. Cell 128: 101–114 [DOI] [PubMed] [Google Scholar]
- Bharadwaj R, Yu H (2004) The spindle checkpoint, aneuploidy, and cancer. Oncogene 23: 2016–2027 [DOI] [PubMed] [Google Scholar]
- Bugreev DV, Yu X, Egelman EH, Mazin AV (2007) Novel pro- and anti-recombination activities of the Bloom's syndrome helicase. Genes Dev 21: 3085–3094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buscaino A, Allshire R, Pidoux A (2010) Building centromeres: home sweet home or a nomadic existence? Curr Opin Genet Dev 20: 118–126 [DOI] [PubMed] [Google Scholar]
- Chaganti RS, Schonberg S, German J (1974) A manyfold increase in sister chromatid exchanges in Bloom's syndrome lymphocytes. Proc Natl Acad Sci USA 71: 4508–4512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KL, North PS, Hickson ID (2007) BLM is required for faithful chromosome segregation and its localization defines a class of ultrafine anaphase bridges. EMBO J 26: 3397–3409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KL, Palmai-Pallag T, Ying S, Hickson ID (2009) Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nat Cell Biol 11: 753–760 [DOI] [PubMed] [Google Scholar]
- Chu WK, Hickson ID (2009) RecQ helicases: multifunctional genome caretakers. Nat Rev Cancer 9: 644–654 [DOI] [PubMed] [Google Scholar]
- Cleveland DW, Mao Y, Sullivan KF (2003) Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell 112: 407–421 [DOI] [PubMed] [Google Scholar]
- Dalal Y, Bui M (2010) Down the rabbit hole of centromere assembly and dynamics. Curr Opin Cell Biol 22: 392–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang G, Yu H, Kirschner MW (1998) Direct binding of CDC20 protein family members activates the anaphase-promoting complex in mitosis and G1. Mol Cell 2: 163–171 [DOI] [PubMed] [Google Scholar]
- Hopfner KP, Michaelis J (2007) Mechanisms of nucleic acid translocases: lessons from structural biology and single-molecule biophysics. Curr Opin Struct Biol 17: 87–95 [DOI] [PubMed] [Google Scholar]
- Hubner NC, Wang LH, Kaulich M, Descombes P, Poser I, Nigg EA (2010) Re-examination of siRNA specificity questions role of PICH and Tao1 in the spindle checkpoint and identifies Mad2 as a sensitive target for small RNAs. Chromosoma 119: 149–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins JR, Toyoda Y, Hegemann B, Poser I, Heriche JK, Sykora MM, Augsburg M, Hudecz O, Buschhorn BA, Bulkescher J, Conrad C, Comartin D, Schleiffer A, Sarov M, Pozniakovsky A, Slabicki MM, Schloissnig S, Steinmacher I, Leuschner M, Ssykor A et al. (2010) Systematic localization and purification of human protein complexes identifies chromosome segregation proteins. Science 328: 593–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima TS, Kawashima SA, Watanabe Y (2004) The conserved kinetochore protein shugoshin protects centromeric cohesion during meiosis. Nature 427: 510–517 [DOI] [PubMed] [Google Scholar]
- Kurasawa Y, Yu-Lee LY (2010) PICH and cotargeted Plk1 coordinately maintain prometaphase chromosome arm architecture. Mol Biol Cell 21: 1188–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng M, Bessuso D, Jung SY, Wang Y, Qin J (2008) Targeting Plk1 to chromosome arms and regulating chromosome compaction by the PICH ATPase. Cell Cycle 7: 1480–1489 [DOI] [PubMed] [Google Scholar]
- Li B, Gogol M, Carey M, Lee D, Seidel C, Workman JL (2007) Combined action of PHD and chromo domains directs the Rpd3S HDAC to transcribed chromatin. Science 316: 1050–1054 [DOI] [PubMed] [Google Scholar]
- Li B, Pattenden SG, Lee D, Gutierrez J, Chen J, Seidel C, Gerton J, Workman JL (2005) Preferential occupancy of histone variant H2AZ at inactive promoters influences local histone modifications and chromatin remodeling. Proc Natl Acad Sci USA 102: 18385–18390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo G, Santoro IM, McDaniel LD, Nishijima I, Mills M, Youssoufian H, Vogel H, Schultz RA, Bradley A (2000) Cancer predisposition caused by elevated mitotic recombination in Bloom mice. Nat Genet 26: 424–429 [DOI] [PubMed] [Google Scholar]
- McGuinness BE, Hirota T, Kudo NR, Peters JM, Nasmyth K (2005) Shugoshin prevents dissociation of cohesin from centromeres during mitosis in vertebrate cells. PLoS Biol 3: e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A, Salmon ED (2007) The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol 8: 379–393 [DOI] [PubMed] [Google Scholar]
- Naim V, Rosselli F (2009) The FANC pathway and BLM collaborate during mitosis to prevent micro-nucleation and chromosome abnormalities. Nat Cell Biol 11: 761–768 [DOI] [PubMed] [Google Scholar]
- Owen-Hughes T, Utley RT, Steger DJ, West JM, John S, Cote J, Havas KM, Workman JL (1999) Analysis of nucleosome disruption by ATP-driven chromatin remodeling complexes. Methods Mol Biol 119: 319–331 [DOI] [PubMed] [Google Scholar]
- Przewloka MR, Glover DM (2009) The kinetochore and the centromere: a working long distance relationship. Annu Rev Genet 43: 439–465 [DOI] [PubMed] [Google Scholar]
- Raynard S, Bussen W, Sung P (2006) A double Holliday junction dissolvasome comprising BLM, topoisomerase IIIalpha, and BLAP75. J Biol Chem 281: 13861–13864 [DOI] [PubMed] [Google Scholar]
- Raynard S, Zhao W, Bussen W, Lu L, Ding YY, Busygina V, Meetei AR, Sung P (2008) Functional role of BLAP75 in BLM-topoisomerase IIIalpha-dependent holliday junction processing. J Biol Chem 283: 15701–15708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosin MP, German J (1985) Evidence for chromosome instability in vivo in Bloom syndrome: increased numbers of micronuclei in exfoliated cells. Hum Genet 71: 187–191 [DOI] [PubMed] [Google Scholar]
- Saha A, Wittmeyer J, Cairns BR (2006) Chromatin remodelling: the industrial revolution of DNA around histones. Nat Rev Mol Cell Biol 7: 437–447 [DOI] [PubMed] [Google Scholar]
- Salic A, Waters JC, Mitchison TJ (2004) Vertebrate shugoshin links sister centromere cohesion and kinetochore microtubule stability in mitosis. Cell 118: 567–578 [DOI] [PubMed] [Google Scholar]
- Singh TR, Ali AM, Busygina V, Raynard S, Fan Q, Du CH, Andreassen PR, Sung P, Meetei AR (2008) BLAP18/RMI2, a novel OB-fold-containing protein, is an essential component of the Bloom helicase-double Holliday junction dissolvasome. Genes Dev 22: 2856–2868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence JM, Phua HH, Mills W, Carpenter AJ, Porter AC, Farr CJ (2007) Depletion of topoisomerase IIalpha leads to shortening of the metaphase interkinetochore distance and abnormal persistence of PICH-coated anaphase threads. J Cell Sci 120: 3952–3964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava V, Modi P, Tripathi V, Mudgal R, De S, Sengupta S (2009) BLM helicase stimulates the ATPase and chromatin-remodeling activities of RAD54. J Cell Sci 122: 3093–3103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z, Bharadwaj R, Li B, Yu H (2001) Mad2-independent inhibition of APCCdc20 by the mitotic checkpoint protein BubR1. Dev Cell 1: 227–237 [DOI] [PubMed] [Google Scholar]
- Tang Z, Shu H, Qi W, Mahmood NA, Mumby MC, Yu H (2006) PP2A is required for centromeric localization of Sgo1 and proper chromosome segregation. Dev Cell 10: 575–585 [DOI] [PubMed] [Google Scholar]
- Tang Z, Sun Y, Harley SE, Zou H, Yu H (2004) Human Bub1 protects centromeric sister-chromatid cohesion through Shugoshin during mitosis. Proc Natl Acad Sci USA 101: 18012–18017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma NH, Czyzewski BK, Alexeev AA, Mazin AV, Kowalczykowski SC, Pavletich NP (2005) Structure of the SWI2/SNF2 chromatin-remodeling domain of eukaryotic Rad54. Nat Struct Mol Biol 12: 350–356 [DOI] [PubMed] [Google Scholar]
- Wang JC (2002) Cellular roles of DNA topoisomerases: a molecular perspective. Nat Rev Mol Cell Biol 3: 430–440 [DOI] [PubMed] [Google Scholar]
- Wang LH, Mayer B, Stemmann O, Nigg EA (2010) Centromere DNA decatenation depends on cohesin removal and is required for mammalian cell division. J Cell Sci 123: 806–813 [DOI] [PubMed] [Google Scholar]
- Wang LH, Schwarzbraun T, Speicher MR, Nigg EA (2008) Persistence of DNA threads in human anaphase cells suggests late completion of sister chromatid decatenation. Chromosoma 117: 123–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y (2005) Shugoshin: guardian spirit at the centromere. Curr Opin Cell Biol 17: 590–595 [DOI] [PubMed] [Google Scholar]
- Wu L, Bachrati CZ, Ou J, Xu C, Yin J, Chang M, Wang W, Li L, Brown GW, Hickson ID (2006) BLAP75/RMI1 promotes the BLM-dependent dissolution of homologous recombination intermediates. Proc Natl Acad Sci USA 103: 4068–4073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Guo R, Sobeck A, Bachrati CZ, Yang J, Enomoto T, Brown GW, Hoatlin ME, Hickson ID, Wang W (2008) RMI, a new OB-fold complex essential for Bloom syndrome protein to maintain genome stability. Genes Dev 22: 2843–2855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagida M (2009) Clearing the way for mitosis: is cohesin a target? Nat Rev Mol Cell Biol 10: 489–496 [DOI] [PubMed] [Google Scholar]
- Yin J, Sobeck A, Xu C, Meetei AR, Hoatlin M, Li L, Wang W (2005) BLAP75, an essential component of Bloom's syndrome protein complexes that maintain genome integrity. EMBO J 24: 1465–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H (2007) Cdc20: a WD40 activator for a cell cycle degradation machine. Mol Cell 27: 3–16 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.