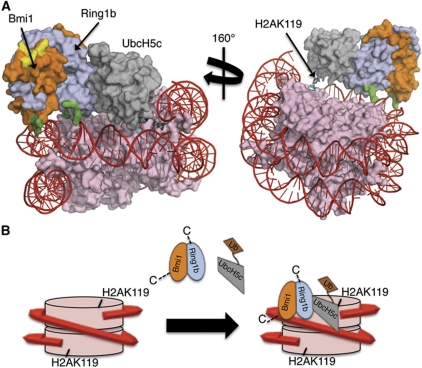
Figure 7.
Model for the interaction of the Bmi1/Ring1b–UbcH5c complex with the nucleosome. (A) Computational docking model generated using HADDOCK2.0. Colour coding is as follows: histones=pink, DNA=red, Ring1b=light blue, Bmi1=orange, UbcH5c=grey. Basic surface residues of Bmi1/Ring1b that were mutated in this study are shaded according to their effect on DNA binding: green=mutation that affected DNA binding, yellow=mutation that had no effect on DNA binding. The C-terminus of histone H2A (cyan stick representation) is the site of ubiquitin modification (K119). UCys85 is coloured by element: nitrogen=blue, oxygen=red, sulphur=yellow. (B) Cartoon model for recognition of the nucleosome by Bmi1/Ring1b–UbcH5c∼Ub complex. Colour scheme is the same as above (ubiquitin is shown in brown). Bmi1/Ring1b uses the basic saddle region to recognize and dock onto the nucleosome through contacts to both DNA and histone H4, positioning UbcH5c∼Ub for ubiquitin transfer to H2AK119. Following transfer of a single ubiquitin, UbcH5c∼Ub is no longer able to access both the E2-binding site on Ring1b and the nucleosome at the same time, leading to termination of the cycle after a single ubiquitination event.