Figure 5. Chromosomal damage as a function of RNAP backtracking.
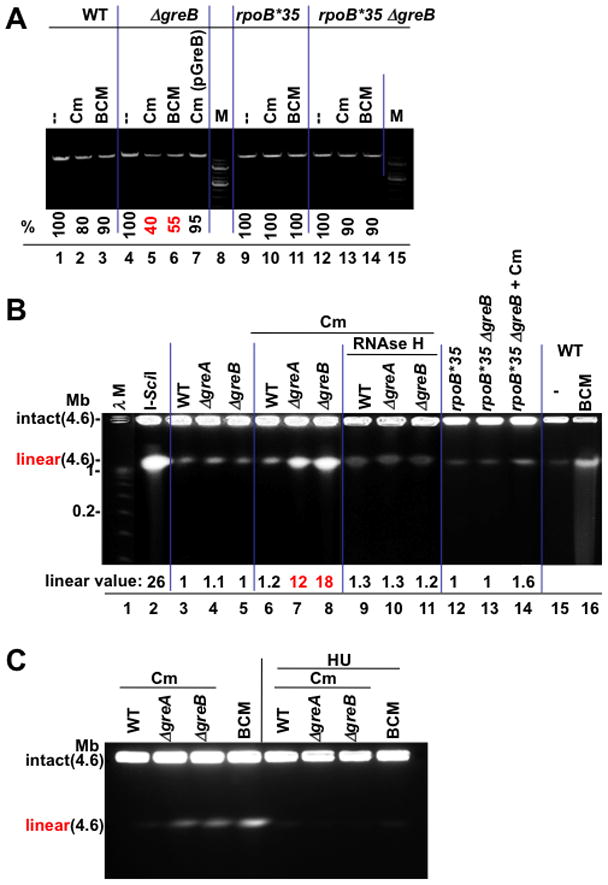
(A) Sub-lethal amounts of the translation inhibitor chloramphenicol (Cm) and BCM produce greater chromosome damage in GreB-deficient cells. The integrity of chromosomal DNA was monitored by PCR. A representative agarose gel shows a 10 kB fragment amplified from equal amounts of genomic DNA isolated from wt (lanes 1–3), ΔgreB (lane 4–6), ΔgreB(pGreB) (lane 7), and rpoB*35 (lanes 9–11), and rpoB*35 ΔgreB (lane 12–14) cells (see Methods). M, 1 kb DNA marker. % indicates the fraction of the full-length PCR products. Values are the average numbers from three experiments with the error margin of less than 5%.
(B) Pulse field gel analysis of chromosomal DSBs; lane 1: λ concatemers from 0.05–1.0 Mb; lane 2: 4.6 Mb linearized E. coli chromosomes (I-SceI); lane 3–5: DNA from wild type (WT) and Gre-deficient cells; lanes 6–8: DNA from wt and Gre-deficient cells after treatment with 4 μg/ml Cm; lanes 9–11: DNA from RNAse H expressing cells after Cm treatment; lanes 12–14: DNA from rpoB*35 and rpoB*35 Gre-deficient cells before and after Cm treatment; lane 16: BCM-treated cells. “Linear value” indicates the fold-increase in linear DNA after Cm or BCM treatment. The values are the average of two or more independent experiments. Note that due to low resolution of PFGE, the species indicated as linear may result from more than one random DSB.
(C) Chromosomal DSBs depend on replication. Pulse field gel analysis of chromosomal DSBs shows that inhibition of replication by HU eliminates all DSBs in wild type and Gre-deficient cells exposed to sub-lethal doses of Cm or BCM.
