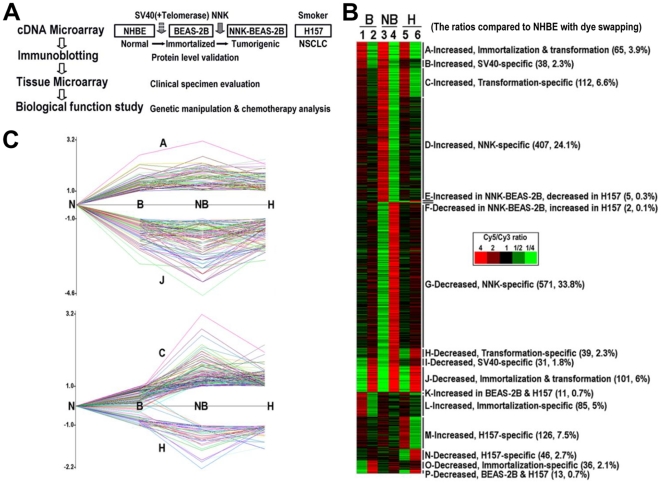
Figure 1. Classification and annotation of differentially expressed genes in BEAS-2B, NNK-BEAS-2B and H157 compared to NHBE.
A. The system and strategy to study gene expression profile, its clinical significance and functional role during NSCLC carcinogenesis and chemotherapeutic resistance. B. Classification of 1,688 regulated genes among BEAS-2B (B), NNK-BEAS-2B (NB) and H157 (H) compared to NHBE (N). Genes whose expression level did not change between cell lines were excluded. The ratios graphically visualized by Cluster and TreeView programs (http://rana.lbl.gov/EisenSoftware.htm). Pairs of lanes and reciprocal red and green colors indicate dye swapping. C. Gene expression dynamics during immortalization and/or transformation. The graphs were generated by Cluster Analysis of Gene Expression Dynamics program (http://www.genomethods.org/caged). A, C, J and H symbols depict groups of genes identified in Figure 1B.