Fig. 2.
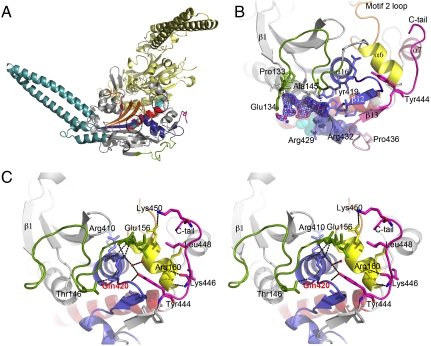
Novel structural elements in C. albicans SerRS_Ser197. (A) Overall structure of the SerRS_Ser197 dimer. Conserved structural elements of monomer A (gray cartoon) are colored in red (motif 1), yellow (motif 2), and purple (motif 3). The N-terminal helical coil, CUG-encoded residues (spheres) at the SerRS dimer interface, and the C-terminal extension characteristic of eukaryotic cytoplasmic SerRSs are colored blue, cyan, and pink, respectively. (B) Secondary nucleotide-binding site at the SerRS surface with bound SerSA (ball-and-stick model with carbon in magenta, oxygen in red, nitrogen in blue, sulfur in orange) and electron density map (1σ contouring, dark blue). The region altered by Ser-to-Leu197 exchange is shown in pale pink. Ser197 (cyan) and Tyr434 (purple) are shown as spheres. (C) Stereo view of the interactions between the Tyr444-centered C-terminal extension (pink) and the SerRS main core. Residues interacting with helices α6, α7, and the α16–β12 loop (sticks), water molecules (red spheres), and putative hydrogen bonds (dashed lines) are shown. The motif 3 helix-capping Gln420 is labeled red.