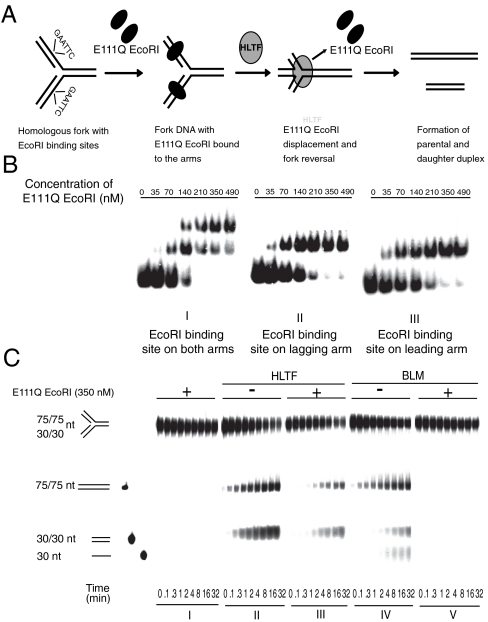
Fig. 1.
Fork reversal activity of HLTF on modeled replication fork bound by dsDNA-binding protein. (A) Schematic representation of a possible mechanism through which HLTF can coordinately remodel a model replication fork bound by E111Q EcoRI. (B) Gel retardation assay showing sequence-specific binding and formation of stable DNA-protein complex by E111Q EcoRI on oligo-based fork-like structures. Increasing amount of E111Q EcoRI was incubated with homologous fork containing an EcoRI binding site. E111Q EcoRI binding to both the arms of the fork is shown in I, whereas II and III show binding to lagging or leading arm only. (C) Comparison of HLTF and BLM fork reversal activities on homologous fork bound by E111Q EcoRI protein on both the arms. In I, control without BLM or HLTF; II, activity of HLTF on naked fork; III, activity of HLTF on E111Q EcoRI-bound fork; IV, activity of BLM on naked fork; V, activity of BLM on E111Q EcoRI-bound fork. Each lane within the panel represents time points at which samples are collected and are noted at the bottom of the gel. Quantitation is shown in Fig. S1.