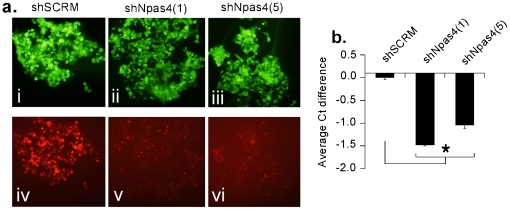
Figure 2. Knockdown of Npas4 in vitro.
(A) HEK293 cells were co-transfected with plasmids expressing Npas4-RFP and plasmids expressing shRNAs designed to deplete Npas4 mRNA [shNpas4(1) (ii, v) and shNpas4(5) (iii, vi)]. A control shRNA (shSCRM) (i, iv) that does not target Npas4 was used as a control. Ninety-six hrs following transfection, cells were visualized by fluorescence microcopy (20X) for the presence of the shRNA GFP plasmids, (panels i, ii, iii) and Npas4-RFP (panels iv, v, and vi). Cells transfected with shNPAS4(1) and shNPAS4(5) exhibited significantly less Npas4-RFP compared to cells transfected with shSCRM, suggesting that the shRNAs against Npas4 are targeting the Npas4-RFP mRNA for degradation resulting in less Npas4-RFP protein (panels v. and vi. versus iv.). Exposure times were the same for visualization of GFP images and RFP images, respectively. (B) qRT-PCR analysis of Npas4 mRNA from samples prepared as described above. Both shNpas4(1) and shNpas4(5) significantly depleted Npas4 mRNA relative to the shSCRM control (n = 3, each group). p<0.05 relative to the shSCRM group.