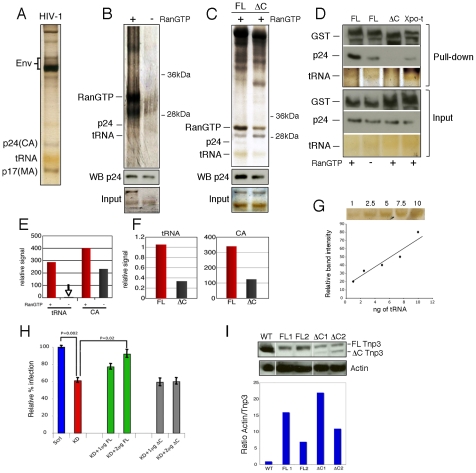
Figure 3. Tnp3 binds viral tRNA and CA.
(A) Purified HIV-1 vector used for pull down assays was visualized by silver staining after 15% denaturing PAGE. Env, viral envelope glycoproteins; p24(CA), capsid proteins; p17(MA), matrix proteins. (B) Pull down assay with purified virus and GST-Tnp3 in the presence or absence of RanQ69L-GTP. (C) Pull down assay with purified virus and GST-Tnp3 (FL) or GST-Tnp3 DC825 (ΔC) deletion mutant in the presence of RanQ69L-GTP. Unlabelled bands are impurities from the Tnp3 preparations. WB p24, Western blot with an anti-capsid antibody. (D) Pull down assay with GST-Tnp3 (FL), GST-Tnp3 DC825 (ΔC) or GST-Xpo-t (Xpo-t). Samples were analyzed by silver stain and by Western blot with anti-capsid and anti-GST antibodies. (E) ImageJ quantification of tRNA and CA in pull down assays as shown in B. RanGTP increased CA binding to Tnp3 by 2.5±0.5 fold in three independent experiments. (F) ImageJ quantification of pull down assays as shown in C. Values are expressed as ratio of input versus recovered tRNAs. DC825 Tnp3 had 3±0.5 fold reduced binding to CA in two independent experiments. (G) tRNA titration curve after silver staining of the SDS-PAGE gel. (H) Rescue experiment. Polyclonal populations of scramble (Scrl) or Tnp3 KD (KD) 293T cells were transfected with plasmid DNA expressing full length (FL) Tnp3 or Tnp3 DC825 bearing a silent point mutation to make them resistant to the shRNA effect (ΔC). Cells were transduced with an HIV-1GFP vector and analyzed by flow cytometry 24 hours later. Mean values ± SD of four independent experiments are shown; Student's t-test was used to calculate statistical significance. (I) Western blot to detect Tnp3 expression in transfected cells. WT, Scrl cells; FL1 and FL2, KD cells transfected with 1 µg and 2 µg Tnp3 cDNA respectively; ΔC1 and ΔC2, KD cells transfected with the same amounts of Tnp3 DC825 cDNA. Bottom panel, quantification of the Tnp3 signal. To quantify signal in FL samples, the background from pre-existing Tnp3 (as detected in the DC825 cDNA-transfected samples) was subtracted and all values were normalized for actin input.