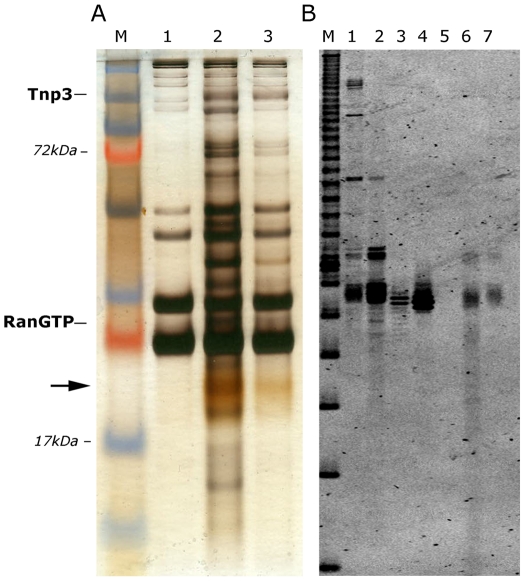
Figure 4. Tnp3 binds cellular tRNAs.
(A) Pull down assays with high-speed cytosolic extracts (HSE) and GST-Tnp3 in the presence or absence of RanQ69L-GTP. Proteins bound to the beads were eluted and analyzed by 15% denaturing SDS-PAGE and silver staining. M, molecular weight protein markers; lane 1, beads only + HSE; lane 2, Tnp3 + HSE + RanQ69L-GTP; lane 3, Tnp3 + HSE without RanQ69L-GTP. The arrow points to the ∼20 kDa tRNA band. (B) Nucleic acids were purified from the same fractions, analyzed by 15% denaturing PAGE and visualized following staining with SYBRgold. Lane M, 10 bp ladder; Lane 1, total RNA from 293T cells; Lane 2, small RNA fraction from 293T cells; Lane 3, in vitro synthesized control tRNA; Lane 4, same tRNA x3; Lane 5, HSE; Lane 6, Tnp3 + HSE + RanQ69LGTP; Lane 7, Tnp3 + HSE without RanQ69LGTP.