Abstract
Klebsiella pneumoniae causes significant morbidity and mortality worldwide, particularly amongst hospitalized individuals. The principle mechanism for pathogenesis in hospital environments involves the formation of biofilms, primarily on implanted medical devices. In this study, we constructed a transposon mutant library in a clinical isolate, K. pneumoniae AJ218, to identify the genes and pathways implicated in biofilm formation. Three mutants severely defective in biofilm formation contained insertions within the mrkABCDF genes encoding the main structural subunit and assembly machinery for type 3 fimbriae. Two other mutants carried insertions within the yfiN and mrkJ genes, which encode GGDEF domain- and EAL domain-containing c-di-GMP turnover enzymes, respectively. The remaining two isolates contained insertions that inactivated the mrkH and mrkI genes, which encode for novel proteins with a c-di-GMP-binding PilZ domain and a LuxR-type transcriptional regulator, respectively. Biochemical and functional assays indicated that the effects of these factors on biofilm formation accompany concomitant changes in type 3 fimbriae expression. We mapped the transcriptional start site of mrkA, demonstrated that MrkH directly activates transcription of the mrkA promoter and showed that MrkH binds strongly to the mrkA regulatory region only in the presence of c-di-GMP. Furthermore, a point mutation in the putative c-di-GMP-binding domain of MrkH completely abolished its function as a transcriptional activator. In vivo analysis of the yfiN and mrkJ genes strongly indicated their c-di-GMP-specific function as diguanylate cyclase and phosphodiesterase, respectively. In addition, in vitro assays showed that purified MrkJ protein has strong c-di-GMP phosphodiesterase activity. These results demonstrate for the first time that c-di-GMP can function as an effector to stimulate the activity of a transcriptional activator, and explain how type 3 fimbriae expression is coordinated with other gene expression programs in K. pneumoniae to promote biofilm formation to implanted medical devices.
Author Summary
Biofilms are surface-associated communities of microorganisms. Biofilm-associated bacteria are protected from host defenses and antibiotics and are the cause of many infections. Klebsiella pneumoniae is primarily a hospital-acquired bacterial pathogen that causes pneumonia, urinary tract infections and septicemia. Its success is related to its ability to form biofilms on medical devices, such as catheters. In K. pneumoniae, biofilm formation is mediated by type 3 fimbriae – hair-like, protein appendages extending out from the cell surface that adhere to surfaces. This study investigated how K. pneumoniae regulates the expression of these fimbriae. We identified a protein, MrkH, which behaves as a “biofilm switch” that turns on the expression of genes responsible for producing type 3 fimbriae. MrkH works by binding to regulatory regions of DNA nearby to these genes and initiates their expression. Importantly, MrkH binds to DNA strongly only when the protein is stimulated by a small molecule, c-di-GMP. Furthermore, we identified bacterial enzymes that either produce or break down c-di-GMP to control its concentration within the cell, and thus modulate MrkH activity. Understanding the molecular basis for these processes may lead to the development of therapeutic compounds, possibly for incorporation into medical device materials to inhibit biofilm formation and pathogenesis.
Introduction
In the first half of the 20th century, Klebsiella pneumoniae was recognized as a community-acquired pulmonary pathogen, chiefly among patients with a history of chronic alcoholism [1]. However, with the advent of more intensive hospital care and the increasingly widespread use of antibiotics, K. pneumoniae has become a significant cause of nosocomially-acquired infections among immunocompromised patients, estimated to cause 8% of all nosocomial infections [2], [3], [4], [5], [6], [7]. These infections commonly include pneumonia, urinary tract infection, septicemia and surgical wound infection.
The pathogenesis of nosocomial K. pneumoniae infections has been associated with its capacity to form biofilms, particularly on medical devices. K. pneumoniae biofilm development is primarily mediated by the mannose-resistant Klebsiella-like (MR/K) hemagglutinins or “Mrk proteins” [8]. The Mrk proteins are encoded by an operon which comprises the genes mrkABCDF [9]. The Mrk proteins form type 3 fimbriae, cell surface structures that can extend into long filaments (up to 2 µm in length) that attach to surfaces [8], [9], [10], [11], [12]. Type 3 fimbriae are synthesized by the chaperone-usher pathway of protein translocation [13]. All chaperone-usher systems are composed of at least three components: (i) a major pilin subunit, polymerized to form the fimbrial shaft, (ii) a chaperone that folds the pilin subunit in the periplasm to ready it for assembly into the fimbriae and (iii) the usher, a transmembrane β-barrel in the outer membrane that polymerizes the pilin subunit into the growing fimbriae, and serves to extrude this growing fimbriae pilin-by-pilin. In Klebsiella, MrkA is the major fimbrial subunit [11], [14], [15] and MrkB and MrkC have the sequence features to represent the periplasmic chaperone and the usher translocase, respectively. The majority of chaperone-usher systems also consist of a fimbria tip-associated adhesion subunit. The MrkD tip adhesion of K. pneumoniae is required for mediating adherence to extracellular matrix proteins such as type V collagen, as well as the basement membrane regions and basolateral surfaces of both renal tract and pulmonary epithelia [10], [12], [14], [16], [17].
An additional complexity for adherence and biofilm formation in K. pneumoniae is the presence of a thick capsule of polysaccharide surrounding the cells. The capsule is important for biofilm formation, since loss-of-function mutations in the cps genes responsible for capsule biosynthesis alter biofilm formation [18], [19]. A previous study, focused on type 1 fimbriae, showed convincingly that fimbria function is inhibited by the presence of the capsule, suggesting direct physical interference with the extension of fimbriae through this 0.5 µm thick, viscous capsule [19]. While it has not been studied, it seems likely that the capsule of K. pneumoniae would also impede extrusion of MR/K hemagglutinins.
As part of a large seroepidemiology study of K. pneumoniae in Australian hospital settings, our laboratory isolated K54-serotype strains at high frequency [20]. These isolates displayed a high degree of clonality, suggesting a common, nosocomial source [20]. The K54 strain AJ218 was found to be significantly more adherent to urinary catheter material and HEp-2 cells than other clinical isolates [20]. These attributes suggest K. pneumoniae AJ218 readily forms biofilms, and may account for the high proportion of infections witnessed in the hospital environment by K54-serotyped strains. We therefore sought to investigate the regulation of biofilm formation in K. pneumoniae AJ218.
Here we report the discovery of a ‘biofilm switch’ controlled by a c-di-GMP-binding protein called MrkH. Our data show that the c-di-GMP-dependent switch between the planktonic and biofilm modes of growth is mediated through control of type 3 fimbriae expression. Both in vivo and in vitro analyses demonstrate that MrkH is a novel PilZ-domain-containing transcriptional regulator, which in the presence of c-di-GMP, activates the expression of the mrkABCDF operon by binding to the region immediately upstream of the mrkA promoter. We also demonstrate that K. pneumoniae encodes a highly active phosphodiesterase (MrkJ) and diguanylate cyclase (YfiN) which appear to contribute to MrkH activity by coordinating cellular c-di-GMP concentration. Together, these results explain how type 3 fimbriae expression in K. pneumoniae is regulated in response to factors signaling for biofilm formation.
Results
Screening of K. pneumoniae transposon mutants for alterations in biofilm formation
To identify factors contributing to rapid biofilm formation by K. pneumoniae AJ218, a 7,000 transposon mutant library was constructed and screened in a polyvinyl-chloride (PVC) microtiter plate assay where biofilm formation was quantified by crystal violet staining [21]. Mutants exhibiting reduced or enhanced biofilm ability greater than 15% of K. pneumoniae AJ218 were then examined individually.
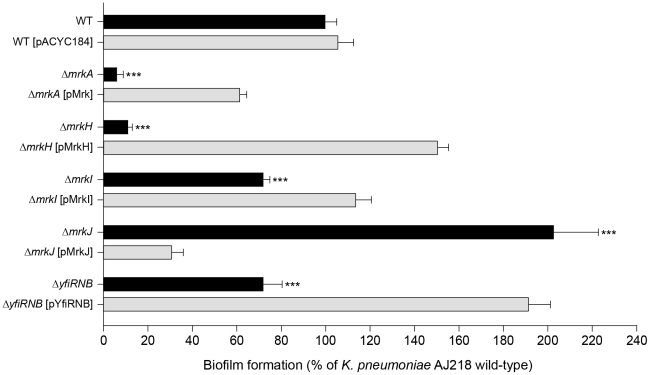
Fifteen biofilm-altered mutants were isolated and the nucleotide sequence immediately flanking each transposon insertion site was identified by Y-linker ligation PCR [22] and sequenced to identify the disrupted locus. Seven mutants were subsequently selected for detailed investigation (Table 1), based upon differences observed in their ability to express functional type 3 fimbriae (see below). The full-length open reading frame (ORF) of each gene disrupted by the transposon insertion was sequenced and exhibited greater than 99% nucleotide identity to homologs located in the sequenced K. pneumoniae KTUH-K2022 genome. To confirm the transposon mutant phenotypes, five deletion mutant strains were constructed from wild-type K. pneumoniae AJ218 whereby the mrkA, mrkH, mrkI, mrkJ and yfiRNB loci were deleted and replaced with a kanamycin resistance-encoding gene. The ΔyfiRNB operon deletion was made to compare to the yfiN transposon mutant, and to analyze mutants that lacked the entire tripartite signalling module. The kanamycin resistance-encoding gene was excised from the ΔmrkH mutant to avoid polar effects on mrkI transcription. Each deletion mutant strain exhibited an equivalent defect in biofilm-formation to the initial transposon mutant (Figure 1). No apparent differences in the planktonic growth rates between wild-type, transposon mutant and deletion mutant strains were observed (data not shown). In complementation analysis, empty vector controls had no impact on biofilm formation for all strains tested (Figure S1).
Table 1. Identification of genetic loci participating in K. pneumoniae AJ218 biofilm formation.
| Strain | Gene name/Locus taga | Predicted protein function and characteristics | Predicted ORF (bp) and protein (aa) length | Biofilm formation of Tn5 mutant (% AJ218Rif)b |
| JW30A8 | mrkA | Major pilin subunit (MrkA) of type 3 fimbriae | 609 bp; 202 aa | 8.9±2.2 |
| KP1_4561 | Main structural component | |||
| JW69A4 | mrkB | Chaperone protein (MrkB) for type 3 fimbriae | 702 bp; 233 aa | 12.7±4.8 |
| KP1_4560 | Assists subunit folding in the periplasm | |||
| JW8E7 | mrkC | Usher protein (MrkC) for type 3 fimbriae | 2487 bp; 828 aa | 13.7±8.5 |
| KP1_4558 | Promotes polymerization of pilin subunits | |||
| JW34H5 | mrkH | C-di-GMP-binding protein (MrkH) | 705 bp; 234 aa | 15.3±6.5 |
| KP1_4551 | Contains PilZ domain | |||
| JW64B4 | mrkI | Transcriptional regulator (MrkI) | 594 bp; 197 aa | 80.5±7.7 |
| KP1_4552 | LuxR superfamily | |||
| Contains HTH DNA-binding motif | ||||
| JW45H9 | mrkJ | Phosphodiesterase (MrkJ) | 717 bp; 238 aa | 248.7±14.4 |
| KP1_4554 | Hydrolyzes c-di-GMP | |||
| Contains EAL domain | ||||
| JW34D8 | yfiN | Diguanylate cyclase (YfiN) | 1224 bp; 407 aa | 86.9±6.1 |
| KP1_4180 | Synthesizes c-di-GMP | |||
| Contains GGDEF domain |
Identification of transposon insertion sites in K. pneumoniae AJ218 were derived from homology searches of the K. pneumoniae NTUH-K2044 genome sequence (GenBank Ref: AP006725).
Expressed as a percentage of the biofilm produced by K. pneumoniae AJ218Rif, set to 100%. Determined using the static microtiter plate assay following incubation in M63B1-GCAA (supplemented with 1% glycerol and 0.3% casamino acids) minimal media for 24 h at 37°C. Values represent the mean ± standard deviation, of four replicate sample wells for each strain performed in two independent experiments.
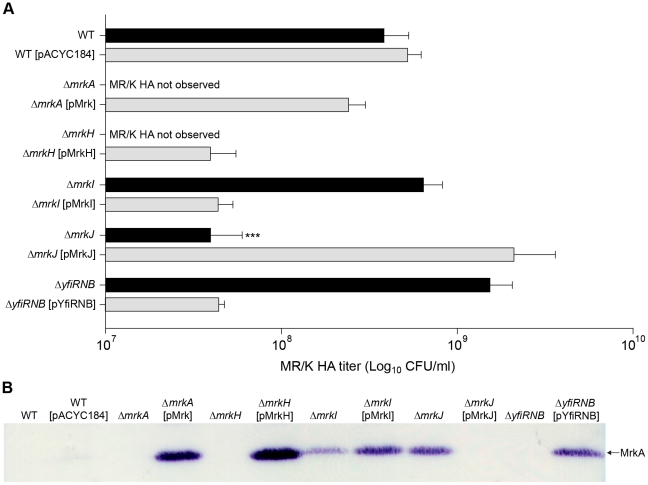
Figure 1. Biofilm formation by K. pneumoniae AJ218.
Biofilm formation by K. pneumoniae AJ218 wild-type, including isogenic mutants and strains harboring trans-complementing plasmids. Biofilm formation was determined using the static microtiter plate assay following incubation in M63B1-GCAA minimal media (supplemented with 1% glycerol and 0.3% casamino acids) for 24 h under static conditions. Results are expressed as a percentage of the biofilm produced by the wild-type AJ218 strain, which is set to 100%. All values represent the mean of four replicate sample wells for each strain performed in two independent experiments. The error bars represent the standard deviation. Statistical significance between AJ218 wild-type and isogenic mutants was analyzed by one-way ANOVA and Tukey HSD post-hoc comparisons are reported, where *** = P<0.001.
Type 3 fimbriae of K. pneumoniae are an important mediator of biofilm formation
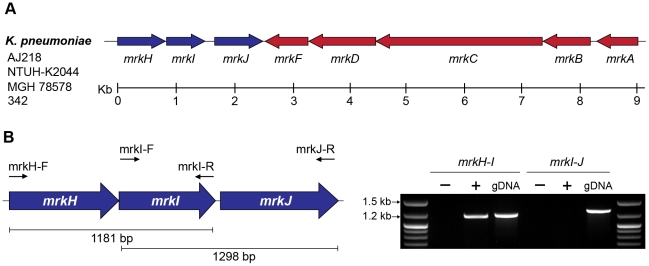
The mrkA and mrkB genes encode the major pilin subunit and periplasmic chaperone of type 3 fimbriae. The mrkC gene encodes a protein of 828 amino acids, predicted to have the three domains defining the usher translocase, where residues 156-651 conform to the central translocase domain as defined by Pfam00577 [23]. In contrast to the K. pneumoniae AJ218 parent strain, any one of the three mutants: ΔmrkA, ΔmrkB or ΔmrkC, failed to form biofilms (Table 1) and promote MR/K hemagglutination, indicating loss of type 3 fimbriae expression. Complete sequencing of the mrk gene cluster of K. pneumoniae AJ218 showed five ORFs (mrkABCDF) arranged in the same transcriptional orientation (Figure 2A), consistent with other K. pneumoniae strains deposited in GenBank.
Figure 2. The mrkABCDF and mrkHIJ loci in K. pneumoniae AJ218.
(A) Genetic organization of the mrkABCDF and mrkHIJ gene clusters from K. pneumoniae strains AJ218, NTUH-K2044 (GenBank Ref: AP006725), MGH 78578 (GenBank Ref: CP000647) and 342 (GenBank Ref: CP000964). (B) RT-PCR analysis of mrkHIJ transcription. PCR amplicon products of either RNA (−), reverse transcribed DNA (+) or genomic DNA (gDNA) were visualized on a 1% agarose gel. The mrkH-I product was generated with primer mrkI-R and amplified with primers mrkH-F and mrkI-R. The mrkI-J product was generated with primer mrkJ-R and amplified with primers mrkI-F and mrkJ-R.
The mrkH, mrkI, mrkJ and yfiRNB loci contribute to K. pneumoniae biofilm formation
Three other mutants isolated from the screen defined a three-locus cluster (mrkH- mrkI-mrkJ) located immediately downstream and transcribed convergently to the mrkABCDF operon (Figure 2A and Table 1). The 9 kb region containing the mrkABCDF and mrkHIJ clusters is highly conserved amongst the sequenced K. pneumoniae genomes, with the nucleotide identity between all strains greater than 99%. Analysis of completed bacterial genome sequences showed that the only other species displaying conserved homologs of these two mrk clusters is Citrobacter koseri, an opportunistic pathogen [24]. Amino acid sequence identities between the Mrk proteins of K. pneumoniae and C. koseri BAA-895 range from 82% (MrkD) to 92% (MrkB), while homology between the mrkHIJ clusters is lower (MrkH = 33%, MrkI = 44%, MrkJ = 73%).
RT-PCR analysis of K. pneumoniae AJ218 cDNA templates spanning intergenic regions between mrkH and mrkI were obtained, but could not be obtained between mrkI and mrkJ (Figure 2B). These results suggest that the mrkH and mrkI genes are co-transcribed in a polycistronic mRNA and mrkJ is transcribed independently of the mrkHI operon.
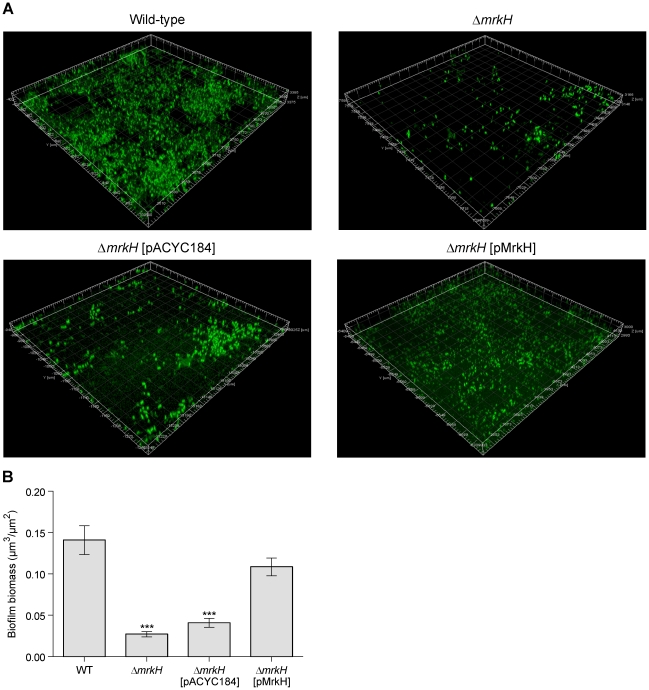
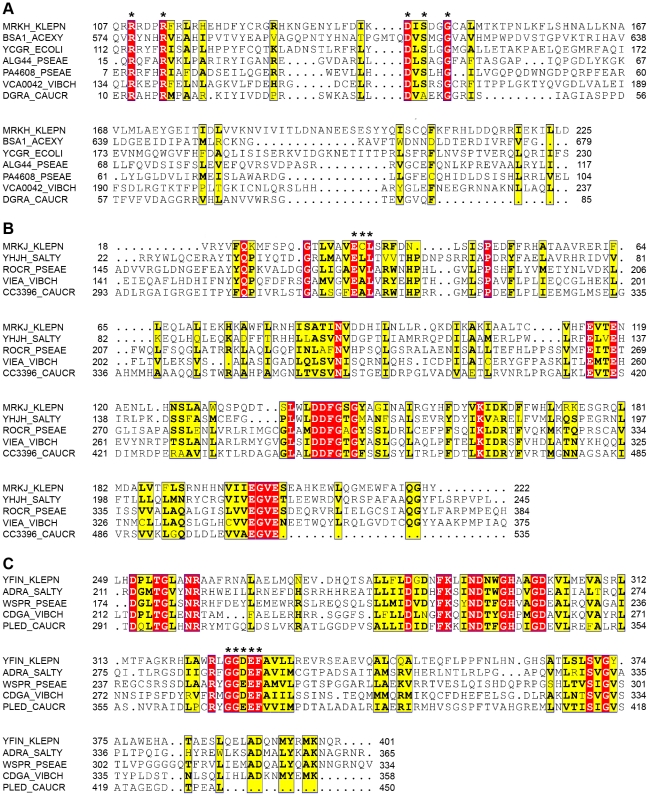
The ΔmrkH mutant was significantly reduced in biofilm formation, both in a static assay that measures initial biofilm formation (Figure 1) and a continuous flow-cell assay that measures mature biofilm development (Figure 3). For both assays, the mutation could be successfully complemented with wild-type mrkH copies (ΔmrkH [pMrkH]). The product of mrkH contains a putative C-terminal PilZ domain capable of binding the second messenger c-di-GMP. Multiple alignments of the PilZ domain of K. pneumoniae MrkH with other experimentally characterized PilZ domain-containing proteins demonstrated complete conservation of two functionally important sequence motifs in MrkH (Figure 4A). The amino acid residues in these conserved motifs (109RxxxR and 140D/NxSxxG) are known to be crucial for c-di-GMP binding and allosteric regulation of PilZ domain-containing proteins for subsequent downstream functions. We therefore propose that MrkH functions as a c-di-GMP-binding protein.
Figure 3. Flow-cell-cultivated biofilm formation by K. pneumoniae AJ218 after 4 days.
(A) Confocal laser scanning micrographs of biofilms formed by K. pneumoniae AJ218 wild-type, ΔmrkH, ΔmrkH [pACYC184] and ΔmrkH [pMrkH] strains. Biofilms were stained with Syto64 to visualize cells and are shown in green as maximum intensity volume rendered projections. (B) COMSTAT analysis of biofilm biomass. The error bars represent the standard deviation. Statistical significance between AJ218 wild-type and ΔmrkH strains was analyzed by one-way ANOVA and Tukey HSD post-hoc comparisons are reported, where *** = P<0.001.
Figure 4. Conservation of PilZ-, EAL- and GGDEF-domain proteins in K. pneumoniae.
Multiple sequence alignment of the (A) PilZ domain of MrkH, (B) EAL domain of MrkJ and (C) GGDEF domain of YfiN from K. pneumoniae AJ218 and other experimentally studied proteins, generated by ClustalW2 [92] and formatted with ESPript [93]. Residues showing strict identity are written in white characters and highlighted in red. Similarity across groups is indicated with black bold characters and highlighted in yellow. Residues required for c-di-GMP binding in the PilZ domain which form two conserved motifs, including the putative catalytic active site residues within the EAL and GGDEF domain, are marked with an asterisk. Protein names and organisms are as follows: MrkH, MrkJ, YfiN: K. pneumoniae AJ218; BcsA: Gluconacetobacter xylinus NBRC 3288, YcgR: E. coli K-12; YhjH, AdrA: Salmonella Typhimurium LT2; Alg44, PA4608, RocR, WspR: Pseudomonas aeruginosa PA14; VCA0042, VieA, CdgA: Vibrio cholerae O395; DgrA, CC3396, PleD: Caulobacter cresentus CB15.
Biofilm formation by the ΔmrkI mutant was moderately deficient compared to wild-type, and could be rescued when supplied with wild-type mrkI copies (ΔmrkI [pMrkI]; Figure 1). The mrkI gene encodes a putative LuxR-like transcriptional regulator that contains a C-terminal helix-turn-helix (HTH) DNA-binding motif. Proteins with LuxR domains can participate in quorum sensing [25], however MrkI lacks the entire N-terminal autoinducer-domain required for binding N-acyl homoserine lactones, suggesting MrkI is a transcriptional activator of gene(s) responding to signals other than those involved in quorum sensing.
The final gene in the three-locus cluster, mrkJ, encodes a putative phosphodiesterase (PDE). This enzyme contains an EAL domain that mediates the hydrolysis of c-di-GMP. Multiple alignments of the EAL domain of MrkJ with other known PDE enzymes demonstrated the conservation of several regions throughout the domain sequence (Figure 4B). Like other enzymes of the same class, the residues ‘ECL’ that form the putative active site of MrkJ varies from the consensus ‘EAL’ sequence. The negative regulatory role of EAL-domain proteins over c-di-GMP levels is consistent with the phenotype of both the ΔmrkJ mutant (enhanced biofilm) and complemented mutant, ΔmrkJ [pMrkJ] (defective biofilm) (Figure 1). This model for biofilm formation is further supported by the identity of the seventh mutant identified in the transposon mutant library screen of K. pneumoniae AJ218 (Table 1). The yfiN gene encodes a putative integral-membrane diguanylate cyclase (DGC) with a GGDEF domain, which functions to synthesize c-di-GMP. Multiple alignment of the GGDEF domain of YfiN with other studied DGC proteins showed conservation of several regions, including the GGDEF residues that form the putative catalytic active site (Figure 4C). Deletion of the entire yfiRNB gene cluster resulted in a significant impairment in biofilm formation and complementation with the yfiRNB operon (ΔyfiRNB [pYfiRNB]) enhanced biofilm formation to levels almost two-fold greater than wild-type (Figure 1).
We therefore propose that biofilm formation in K. pneumoniae is regulated by the relative availability of intracellular c-di-GMP, coordinated by c-di-GMP turnover enzymes (YfiN and MrkJ), and sensed by a receptor protein (MrkH). When a PDE is inactivated or a DGC up-regulated leading to increased c-di-GMP availability, biofilm formation is stimulated. Conversely, when a DGC is inactivated or a PDE up-regulated resulting in decreased c-di-GMP concentration, biofilm formation is inhibited.
MrkH positively regulates type 3 fimbriae expression in K. pneumoniae
We sought to address whether the activity of MrkH influences type 3 fimbriae expression. A sensitive functional assay for type 3 fimbriae is the MR/K hemagglutination (HA) activity, mediated by the MrkD adhesion located at the tips of type 3 fimbriae. As expected, the ΔmrkA mutant failed to mediate a visible MR/K HA reaction at the highest bacterial density tested (approximately 1×1010 CFU/mL; Figure 5A). Likewise, the ΔmrkH mutant completely lacked MR/K HA activity at the same cell concentration. Upon complementation, strong MR/K HA activity was observed for both strains. The MR/K HA titer for the complemented strains was either reduced to wild-type level (for the ΔmrkA-complemented mutant) or to approximately six-fold lower than wild-type level (for the ΔmrkH-complemented mutant). It was shown that empty vector controls had no impact on MR/K HA expression for all strains tested (Figure S2). A complementary assay for MR/K HA expression utilized a specific anti-MrkA antibody in immunoblot analysis of total cellular extracts. As expected, MrkA could not be detected in the ΔmrkA mutant, while expression was restored when the mutant was complemented with the complete mrkABCDF gene cluster (ΔmrkA [pMrk]; Figure 5B). Using this assay, MrkA was not detected in wild-type K. pneumoniae AJ218, indicating that this strain only weakly expresses type 3 fimbriae in planktonic culture. Similarly, MrkA was not detected in the ΔmrkH mutant, but was expressed above wild-type levels when the mutant was complemented with a plasmid expression construct for MrkH (ΔmrkH [pMrkH]; Figure 5B). Therefore, when MrkH is over-expressed in K. pneumoniae, both the production of the MrkA subunit and the MR/K HA activity is increased. MrkH is therefore a critical, positive regulator of type 3 fimbriae expression.
Figure 5. Type 3 fimbriae expression by K. pneumoniae AJ218.
(A) Mannose resistant Klebsiella-like hemagglutination (MR/K HA) assays using human erythrocytes. MR/K HA titer is expressed as the lowest concentration (CFU/mL) of bacteria causing a visible agglutination reaction. Values represent the mean of three independent experiments. The error bars represent the standard deviation. Statistical significance between AJ218 wild-type and isogenic mutants was analyzed by one-way ANOVA and Tukey HSD post-hoc comparisons are reported, where *** = P<0.001. (B) Cell lysates were prepared from the indicated strains and analyzed by SDS-PAGE and immunoblotting with anti-MrkA antiserum. The MrkA pilin monomer (which migrates at approximately 21 kDa) is labeled.
Regulation of type 3 fimbriae expression in K. pneumoniae by YfiRNB and MrkJ
We also examined the levels of MrkA subunit production and MR/K HA activity of ΔyfiRNB (DGC) and ΔmrkJ (PDE) mutant strains (Figure 5B). The MrkA subunit was not detected in the ΔyfiRNB mutant, but steady-state levels of MrkA were higher than wild-type in mutants complemented with the wild-type yfiRNB operon. Conversely, MrkA levels were increased in the ΔmrkJ mutant, but absent when complemented with wild-type mrkJ gene copies. The MR/K HA activity of these strains was consistent with the immunoblot results (Figure 5A). These observations suggest that YfiRNB and MrkJ function, respectively, as positive and negative regulators of type 3 fimbriae expression. The phenotypes of the mutants are consistent with their roles in modulating the intracellular levels of c-di-GMP.
MrkI, the LuxR-like regulator, mediates type 3 fimbriae functionality in K. pneumoniae
The hemagglutination tests demonstrated that the ΔmrkI mutant, shown previously to be deficient in biofilm formation, also had decreased MR/K HA activity. This is consistent with a reduced amount of type 3 fimbriae on the cell surface. These results suggest that MrkI functions as a positive regulator of type 3 fimbriae. Counter-intuitively, the ΔmrkI mutant appeared to express more MrkA subunit than the wild-type strain. Moreover, when complemented, the over-expressed MrkI strain appeared to produce even greater amounts of MrkA subunit. Why would the increased levels of MrkA produced in the ΔmrkI mutant not be assembled into functional fimbriae to facilitate biofilm formation and MR/K HA?
We hypothesize that this discrepancy is due to MrkI exhibiting multiple roles within the cell. It could function as a minor transcriptional activator of type 3 fimbriae synthesis, in conjunction with a role to regulate another component of the fimbriae assembly pathway (e.g., the usher translocase, MrkB). Alternatively, MrkI could regulate other cell surface factors, such as the polysaccharide capsule, which may in turn affect the function of type 3 fimbriae, as previously described for type 1 fimbriae [19].
In this scenario, a decrease in type 3 fimbriae assembly when MrkI is absent could lead to intracellular accumulation of MrkA, seen as an apparent increase in the steady-state level of MrkA by immunoblot (Figure 5B), resulting in the observed deficiency in MR/K HA and biofilm formation. When MrkI is over-expressed, the functionality of MrkA is restored and type 3 fimbriae expression and biofilm formation is enhanced greater than wild-type levels. The increased levels of MrkA subunit produced in the ΔmrkI complemented strain is suggestive of MrkI functioning as a strong activator of mrkA transcription when present in high numbers.
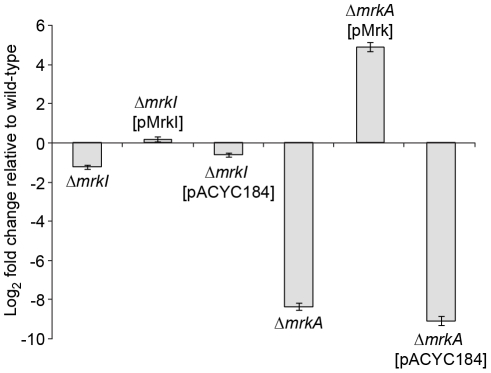
Using real-time PCR, only a slight decrease in mrkA gene transcript expression is observed in the ΔmrkI mutant, consistent with MrkI having a partial role in type 3 fimbriae activation (Figure 6). However, we failed to see a dramatic increase in mrkA RNA levels in the ΔmrkI complemented strain that could account for the large elevation in MrkA protein production seen in the cell extracts. That MrkB, the periplasmic chaperone, and MrkC, the usher translocase, are required to assemble and export the MrkA protein might account for this. However, we cannot rule out that MrkI also positively regulates other gene(s) that encode factors which participate in the correct folding of the MrkA fimbrial subunit.
Figure 6. Quantitative RT-PCR analysis of mrkA RNA levels.
Fold differences in mrkA transcript expression levels compared to K. pneumoniae AJ218 wild-type levels are shown for the indicated K. pneumoniae strains. The mrkA transcription expression was normalized to rpoD concentrations. Values represent the mean of reactions performed in triplicate. The error bars represent the standard deviation.
MrkH positively controls the transcription of the mrkA promoter
We sought to test whether MrkH activates transcription of the mrkABCDF operon by stimulating a promoter(s) located in the upstream region of this gene cluster. The sequence of this upstream region is shown in Figure 7A. To test this hypothesis, we established an assay system in Escherichia coli using a lacZ reporter positioned downstream of specific regulatory regions of the mrkA gene (Materials and Methods). Initially, two mrkA-lacZ fusions were constructed: mrkA-lacZ-1 and mrkA-lacZ-2, spanning positions −759 to +109 (mrkA-lacZ-1) and from −295 to +109 (mrkA-lacZ-2) relative to the mrkA translational start codon. These two plasmids, along with the “promoterless” control plasmid pMU2385, were each transformed into E. coli K12 strain MC4100 (ΔlacZ) containing either vector pACYC184 (MrkH–) or its derivative pMrkH (MrkH+) and assayed for β-galactosidase expression.
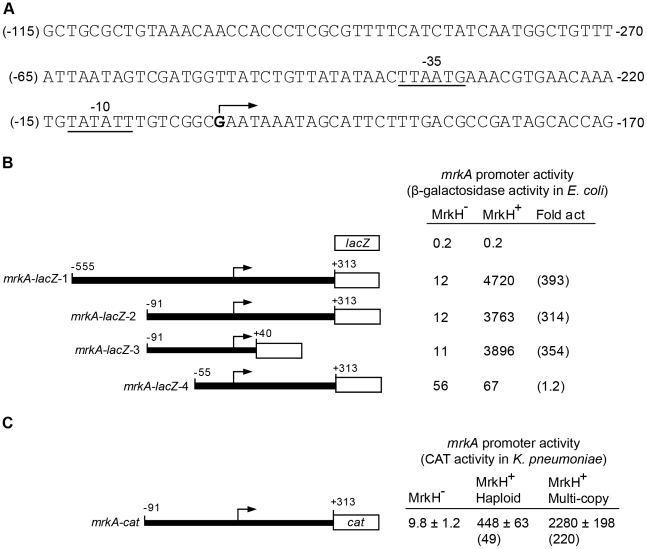
Figure 7. Transcriptional analysis of the mrkA regulatory region.
(A) The nucleotide sequence of the mrkA regulatory region is shown. The numbering on the left of the sequence (in brackets) is relative to the transcriptional start site of mrkA. The numbering on the right of the sequence is relative to the start codon of the mrkA coding sequence. The transcriptional start site is marked with an angled arrow and the putative -35 and -10 regions of the mrkA promoter are indicated and underlined. (B) Promoter activities of various mrkA-lacZ transcriptional fusions in the MrkH– (MC4100 containing pACYC184) and MrkH+ (MC4100 containing pMrkH) E. coli backgrounds are shown as specific activities of β-galactosidase (Miller) units which are the mean values from three independent assays, with variation <15%. Fold activation (Fold act.) is the specific activity of β-galactosidase of the MrkH+ strain divided by that of the MrkH− strain. The numbers shown above the various mrkA fragments are relative to the start site of transcription (angled arrows) and the lengths of the various mrkA fragments are not to scale. (C) The effect of MrkH on transcription of the mrkA promoter was analyzed by a CAT assay in three isogenic K. pneumoniae backgrounds: MrkH− (ΔmrkH + pACYC184-KmR), MrkH+ haploid (wild-type + pACYC184-KmR), and multi-copy MrkH+ (ΔmrkH + pACYC184-KmR-mrkH). Specific CAT activities are the averages of three independent assays and the standard deviation values are shown. Values in brackets are fold activation (for details, see above).
Compared to the negative control (pMU2385), both mrkA-lacZ-1 and mrkA-lacZ-2 produced low but significant levels of β-galactosidase activity (12 U) in the MrkH– background (Figure 7B). In both cases, MrkH stimulated expression of mrkA-lacZ-1 and mrkA-lacZ-2 more than 300-fold. These results demonstrate the presence of a promoter(s) in the upstream region of mrkA, which is highly activated by MrkH. Given the similar regulatory patterns of the two constructs, both the promoter and operator elements are present within the 404 bp fragment carried on mrkA-lacZ-2.
To verify MrkH-mediated activation of mrkA transcription directly in K. pneumoniae, the 404 bp mrkA fragment (as carried by mrkA-lacZ-2) was inserted upstream of the cat reporter gene in plasmid pKK232-8. The resulting plasmid, mrkA-cat, was then introduced into three isogenic K. pneumoniae strains: the wild-type AJ218 strain (MrkH+ haploid), the ΔmrkH mutant (MrkH–) and wild-type AJ218 carrying pMrkH (multi-copy MrkH+). As expected, ΔmrkH mutants express barely-detectable CAT activity (9.8 U; Figure 7C). In contrast, when K. pneumoniae carried the wild-type mrkH gene, CAT expression increased 49-fold to 448 U. Further evidence of the stimulatory role of MrkH on transcriptional activation comes from the CAT expression measured (2000 U) when multiple copies of the mrkH gene are present.
Mapping the mrkA transcriptional start site and the region required for MrkH activation
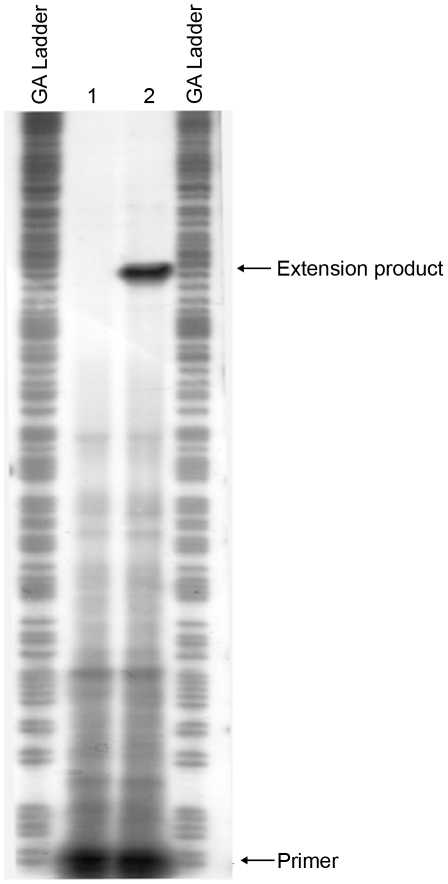
To map the transcriptional start site(s) of mrkA, we performed a primer extension experiment. Total cellular RNA was isolated from E. coli MC4100 strains containing pMrkH with either pMU2385 (control) or mrkA-lacZ-2. Following hybridization of the RNA with 32P-labelled primer (Px1mrkARev) and extension with CMV reverse transcriptase in the presence of dNTPs, the samples were analyzed on a sequencing gel. A single extension product was evident from the mrkA-lacZ-2 sample (Figure 8). The data mapped the start site of transcription to 204 bp upstream of the putative start site of translation of mrkA (Figure 7A). Inspection of the sequence revealed the presence of the hexanucleotides TATATT centered at -10.5 relative to the start site of transcription, which is a good match to the consensus sequence of the -10 region of a bacterial σ70 promoter. The only possible -35 sequence (TTAATG), which matches poorly to the consensus sequence, was found 15 bp upstream of the putative -10 region. The combination of a poor -35 region and imperfect spacing may contribute to the very weak promoter activity observed in the MrkH− background.
Figure 8. Mapping the start site of transcription of the mrkA promoter by primer extension.
Total cellular RNA was purified from E. coli MC4100 strains containing pMrkH with either pMU2385 (control) or mrkA-lacZ-2. The RNA samples were then hybridized with 32P-labelled primer Px1mrkARev. Primer extension was performed using AMV reverse transcriptase in the presence of dNTPs. GA Ladder: GA sequence ladder prepared using the mrkA PCR fragment generated using primer pairs 32P-Px1mrkARev and mrk295F. Lane 1: control experiment using RNA from E. coli MC4100 strain containing pMrkH and pMU2385. Lane 2: experiment using RNA from E. coli MC4100 strain containing pMrkH and mrkA-lacZ-2. The positions corresponding to 32P-Px1mrkARev primer and the extension product are marked.
Mapping the mrkA promoter allowed us to make more precise deletion constructs: mrkA-lacZ-3 and mrkA-lacZ-4, in order to localize the region responsible for MrkH-mediated activation of mrkA transcription. As shown in Figure 7B, construct mrkA-lacZ-3, in which most nucleotides downstream of the start site of transcription were deleted, exhibited the same degree of activation by MrkH as mrkA-lacZ-2, indicating that the deleted downstream sequence (between +40 and +313) was not required for MrkH activation. In the case of mrkA-lacZ-4, however, removal of the upstream region between -91 and -56 caused a 5-fold increase in basal level promoter activity in the MrkH− background and completely abolished the MrkH-mediated transcriptional activation of the mrkA promoter in the MrkH+ background (Figure 7B). From these results, we determined that a cis-acting element responsible for MrkH-mediated activation was located between positions -91 and -56.
C-di-GMP facilitates binding of MrkH to the mrkA regulatory region
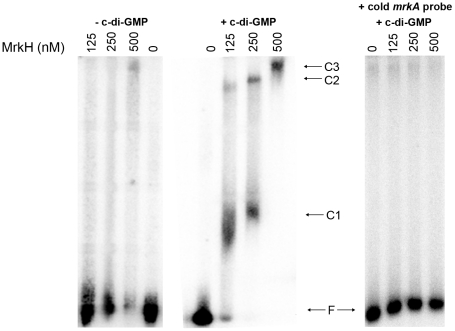
To test whether MrkH was able to bind directly to the mrkA regulatory region, we expressed and purified recombinant MrkH (MrkH-8×His) and used it in an electrophoretic mobility gel shift assay (EMSA). The purity of the MrkH-8×His preparation is shown in Figure S3. The mrkA fragment which spanned between −91 and +160, relative to the start site of transcription, was end-labeled with 32P and incubated with varying amounts of MrkH-8×His in the absence or presence of 200 µM c-di-GMP at 37°C for 20 min. The samples were then analyzed on native polyacrylamide gels. In the absence of c-di-GMP, no shift of DNA was seen at the MrkH-8×His concentration of 125 nM (Figure 9). Increasing the protein concentration to 250 or 500 nM resulted in the partial shift of DNA; however, no discrete protein-DNA band was obvious. In the presence of c-di-GMP, the majority of the DNA was shifted to form a major protein-DNA complex (C1) and a minor complex (C2) at the MrkH-8×His concentration of 125 nM. At the higher protein concentration of 250 nM, the intensity of the larger C2 complex was enhanced. Increasing the protein concentration to 500 nM led to a complete shift of DNA and the formation of a single, even larger protein-DNA complex (C3; Figure 9). The right panel of Figure 9 showed that the addition of specific cold competitor DNA can out-compete the binding of MrkH to the labeled probe, demonstrating that MrkH binds specifically to the mrkA promoter region. Two additional control experiments were carried out in which the mrkA promoter fragment was incubated with either the wild-type MrkH-8×His in the presence of GTP or the mutant MrkH-8×His (113R-A, for details see below) in the presence of c-di-GMP. EMSA analysis showed that under these conditions neither the wild-type nor the mutant MrkH can form a protein-DNA complex with the mrkA promoter fragment (Figure S4).
Figure 9. Analysis of the binding of MrkH-8×His to the mrkA regulatory region by EMSA.
The 32P-labelled PCR fragment containing the mrkA regulatory region was generated using primer pairs 32P-Px1mrkARev and mrk295F. The mrkA fragment was mixed with varying amounts of the purified MrkH-8×His protein (from 0 to 500 nM) in the absence or presence of c-di-GMP (200 µM). Following incubation at 30°C for 20 min, the samples were analyzed on native polyacrylamide gels. The right-hand panel shows control reactions with approximately 100-fold molar excess of the unlabeled (cold) mrkA promoter fragment (specific competitor DNA), used to demonstrate the specificity of the c-di-GMP-mediated MrkH binding to the mrkA promoter region. The unbound DNA (F) and protein-DNA complexes (C1, C2 and C3) are marked.
The results of EMSA demonstrated that (i) MrkH is a DNA binding protein, (ii) c-di-GMP facilitates the binding of MrkH to the mrkA regulatory region and (iii) MrkH can oligomerise on DNA to form a very large MrkH:DNA complex.
C-di-GMP positively controls the activity of the MrkH protein
To provide further evidence that c-di-GMP positively regulates MrkH function, we performed mutational analysis of the mrkH gene. Construct mrkH:113R-A contained a point mutation in which the conserved arginine residue at position 113 within the putative c-di-GMP-binding PilZ domain was replaced with an alanine residue (Figures 4A and 10A). An immunoblot of a C-terminal His8-tag fusion with MrkH:113R-A demonstrated that this mutant construct was stably expressed in E. coli MC4100 (Figure S5).
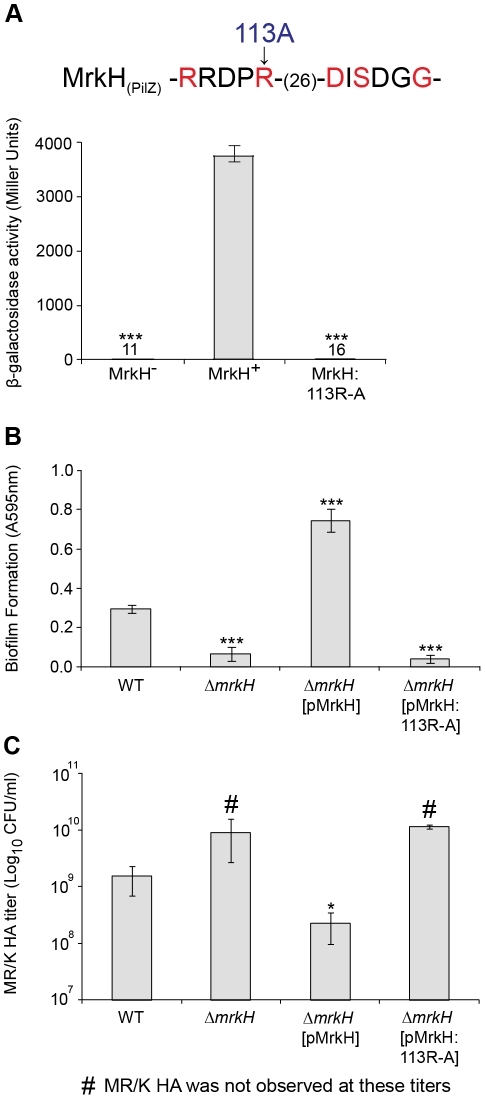
Figure 10. Analysis of a PilZ-domain mutation on MrkH-mediated transcriptional activation, biofilm formation and type 3 fimbriae expression.
(A) Amino acids within the conserved PilZ domain are shown and residues in red are those known to be critical for c-di-GMP binding in other studies [50]. As indicated, the MrkH mutation carries an arginine to alanine change at position 113. β-galactosidase assays were performed with E. coli MC4100 strains: MrkH− (carrying reporter plasmid mrkA-lacZ-2 and pACYC184), MrkH+ (carrying mrkA-lacZ-2 and pMrkH) and MrkH:113R-A (carrying mrkA-lacZ-2 and pMrkH:113R-A). The specific β-galactosidase activities are as follows: MrkH−: 11±0.9; MrkH+: 3760±186 and MrkH:113R-A: 16±1.5. Values represent the mean of three replicate samples. (B) Static biofilm formation assay by the indicated K. pneumoniae strains. Values represent the mean of four replicate sample wells for each strain performed in two independent experiments. (C) MR/K HA assay of the indicated K. pneumoniae strains. Values represent the mean of three independent experiments. The error bars represent the standard deviation. Statistical significance between MrkH+ and other strains (β-galactosidase assay) was analyzed by the Van der Waerden test, and significance between AJ218 wild-type and ΔmrkH strains (biofilm and MR/K HA assays) was analyzed by one-way ANOVA. Tukey HSD post-hoc comparisons are reported, where *** = P<0.001, * = P<0.05.
To examine the ability of MrkH:113R-A to activate the transcription of mrkA, pACYC184 carrying the mrkH:113R-A mutation, along with pACYC184 carrying wild-type mrkH (MrkH+) and pACYC184 alone (MrkH−), were transformed into E. coli MC4100 containing the single-copy reporter plasmid mrkA-lacZ-2. These strains were then assayed for β-galactosidase activity following mid-log growth in LB media. As expected, the wild-type MrkH+ exerted more than 300-fold activation of the mrkA promoter (Figure 10A). Consistent with our prediction that MrkH is essential for c-di-GMP-mediated positive control of mrkA transcription, the substitution mutation within the PilZ domain completely destroyed the ability of MrkH to activate transcription from the mrkA promoter.
To examine the effect of the PilZ-domain mutation on biofilm formation and type 3 fimbriae expression of K. pneumoniae AJ218, we introduced the mrkH:113R-A construct described above into the ΔmrkH mutant and assayed the resulting strain in static biofilm formation and MR/K HA activity. Although strong biofilm formation (Figure 10B) and MR/K HA activity (Figure 10C) was observed by the ΔmrkH mutant upon complementation with wild-type mrkH, the mrkH:113R-A construct was completely ineffective and failed to rescue the ΔmrkH mutant phenotypes.
These results strongly indicated that the c-di-GMP binding region within the PilZ domain of MrkH is critical for transcriptional activation of the mrk operon and subsequent biofilm formation via type 3 fimbriae expression.
MrkH activity is influenced by MrkJ- and YfiN-mediated control of c-di-GMP expression levels
If the above conclusion is correct, we would expect that a decrease in intracellular c-di-GMP levels by enhanced expression of a phosphodiesterase causes a reduction in MrkH activity. Conversely, increasing the endogenous c-di-GMP concentration by enhancing the expression of a diguanylate cyclase would lead to greater MrkH activity.
To test this hypothesis, we introduced pBR322 derivatives carrying either the wild-type mrkJ gene, the yfiRNB operon, or pBR322 alone into E. coli MC4100, which contained pACYC184 carrying wild-type mrkH (MrkH+) and the reporter plasmid mrkA-lacZ-2. To confirm the roles of MrkJ and YfiN as c-di-GMP-specific phosphodiesterase and diguanylate cyclase, respectively, we also generated site-directed mutant constructs of these enzymes in their conserved c-di-GMP hydrolysis/synthesis catalytic sites and tested their activity alongside the wild-type constructs. Construct mrkJ:36ECL-AAA contained substitution mutations in which the EAL domain of MrkJ (starting at residue 36) was replaced with alanine residues (Figures 4B and 11A). In addition, construct yfiRNB:328DEF-AAA carried substitution mutations in which the DEF residues of the GGDEF domain of YfiN (starting at residue 328) were replaced with alanine residues (Figures 4C and 11A).
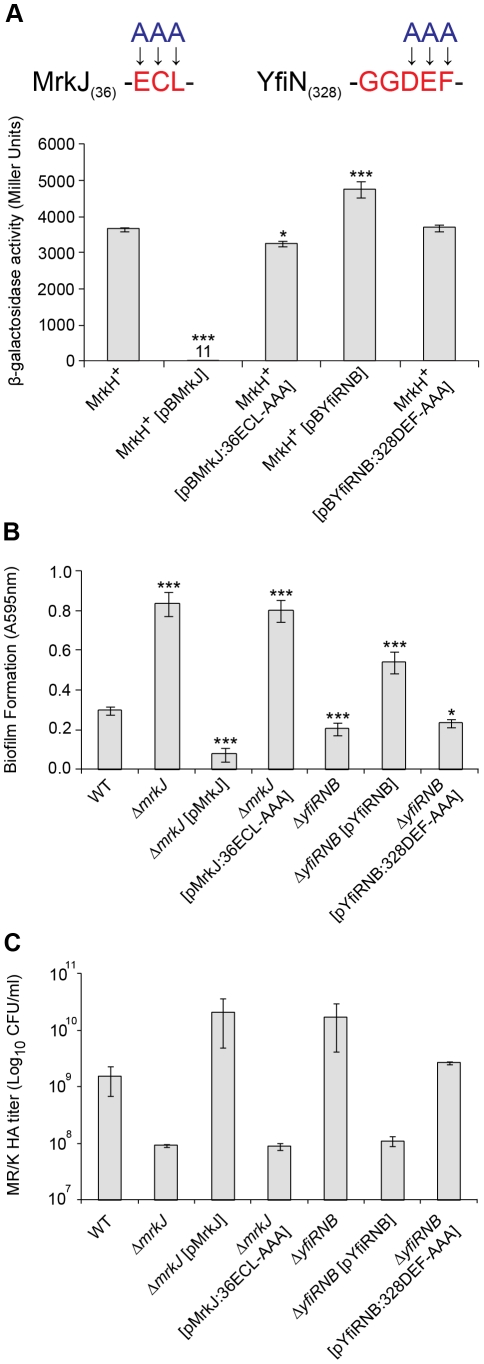
Figure 11. Analysis of EAL- and GGDEF-domain mutations on MrkH-mediated transcriptional activation, biofilm formation and type 3 fimbriae expression.
(A) Amino acids within the conserved EAL and GGDEF domains of MrkJ and YfiN were substituted with alanine residues. β-galactosidase assays were performed with: E. coli MC4100 strain MrkH+ (carrying reporter plasmid mrkA-lacZ-2 and pMrkH), including pBR322-derived vectors pBMrkJ (wild-type), pBMrkJ:36ECL-AAA, pBYfiRNB (wild-type) or pBYfiRNB:328DEF-AAA. Values represent the mean of three replicate samples. (B) Static biofilm formation assay by the indicated K. pneumoniae strains. Values represent the mean of four replicate sample wells for each strain performed in two independent experiments. (C) MR/K HA assay of the indicated K. pneumoniae strains. Values represent the mean of three independent experiments. The error bars represent the standard deviation. Statistical significance between MrkH+ (wild-type) and other strains (β-galactosidase assay), as well as AJ218 wild-type and isogenic mutant strains (MR/K HA assay) was analyzed by the Van der Waerden test. Significance between AJ218 wild-type and isogenic mutant strains (biofilm assay) was analyzed by one-way ANOVA. Tukey HSD post-hoc comparisons are reported, where *** = P<0.001, * = P<0.05.
These strains were then assayed for β-galactosidase activity following mid-log growth in LB media. Enhanced expression of wild-type MrkJ led to a complete inability of MrkH to activate transcription from the mrkA promoter (Figure 11A). Conversely, mutation of the EAL domain of MrkJ led to enhanced MrkH-mediated transcriptional activation. This result suggested that an inactive EAL domain renders MrkJ unable to effectively hydrolyze c-di-GMP, permitting c-di-GMP accumulation and increased MrkH activity. The increased expression of wild-type YfiRNB resulted in a small but significant enhancement in MrkH-mediated activation of mrkA transcription. However, this increase was not observed from the YfiN construct containing the mutated GGDEF domain, suggesting that the lowered MrkH-mediated activation of the mrkA promoter was caused by YfiN impairment in c-di-GMP synthesis. The site-directed mutations of the EAL and GGDEF domains of MrkJ and YfiN, respectively, also had pronounced effects on biofilm formation and type 3 fimbriae synthesis by K. pneumoniae AJ218. The mrkJ:36ECL-AAA and yfiRNB:328DEF-AAA gene constructs also failed to effectively complement the respective ΔmrkJ and ΔyfiRNB mutant strains in static biofilm formation (Figure 11B) and MR/K HA activity (Figure 11C).
Taken together, these results indicate that MrkJ and YfiN are c-di-GMP-specific phosphodiesterase and diguanylate cyclase, respectively. Moreover, we have clearly demonstrated that c-di-GMP is a positive cofactor essential for MrkH-mediated transcriptional activation of the mrk operon for type 3 fimbriae synthesis and biofilm formation by K. pneumoniae.
Detection of phosphodiesterase activity for the MrkJ protein
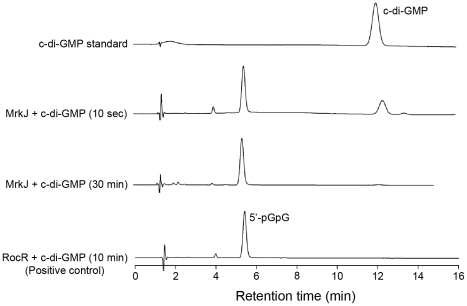
From the data presented, MrkJ represents a c-di-GMP-specific phosphodiesterase that has pronounced negative effects on K. pneumoniae biofilm formation. To further characterize the kinetics of c-di-GMP hydrolysis by MrkJ, we expressed and purified MrkJ (MrkJ-8×His) and analyzed its enzyme activity by High-Performance Liquid Chromatography (HPLC). The purity of the MrkJ-8×His preparation is shown in Figure S6. We demonstrated that a significant amount of c-di-GMP was hydrolyzed by MrkJ-8×His to form the degradation product 5′-pGpG after 10 sec (Figure 12). The c-di-GMP was completely converted to 5′-pGpG after 30 min. These results confirmed that MrkJ possesses very strong phosphodiesterase activity, a significant feature for an inhibitor of biofilm formation.
Figure 12. MrkJ displays strong phosphodiesterase activity.
C-di-GMP hydrolysis by purified MrkJ was analyzed by HPLC. The formation of 5′-pGpG was monitored at various time-points and the c-di-GMP-specific phosphodiesterase RocR [94] was used as the positive control.
Discussion
The role of type 3 fimbriae in K. pneumoniae
The type 3 fimbriae of K. pneumoniae, encoded by the mrkABCDF operon, represent a surface appendage that is significant in mediating biofilm formation. As the major pilin, MrkA is required to extend the fimbriae [8], [9], which is particularly important in the case of K. pneumoniae given the thick capsule surrounding the outer membrane [19]. Past studies have shown that ΔmrkA mutants have a greatly diminished capacity for adherence to abiotic surfaces and for biofilm formation in continuous flow-through chambers [14], [15], [26]. We found that during planktonic growth very little MrkA is produced, with only a sensitive functional assay able to detect fimbria-dependent hemagglutination. However, the type 3 fimbriae are essential for biofilm growth of K. pneumoniae AJ218 in flow-cell chambers and adherence to PVC surfaces, a common material used in indwelling medical devices, such as catheters and endotracheal tubes.
Type 3 fimbriae are found in several groups of Enterobacteriaceae [27], and are extended by a step-wise addition of MrkA subunits to the base of the filament [13], [28]. Each MrkA subunit is taken up by the N-terminal domain of the Usher translocase, with the translocase thereby catalyzing polymerization and providing the pore through which the filament passes across the outer membrane. In K. pneumoniae, these fimbriae must extend from the outer membrane through a thick carbohydrate capsule to reach, and extend beyond, the actual cell surface. Comparative genomics and experimental evidence suggests that most clinical and environmental strains of K. pneumoniae carry the genes required to produce type 3 fimbriae [29], [30]. The K. pneumoniae AJ218 mrkABCDF operon is almost identical in nucleotide sequence to other K. pneumoniae clinical strains. This suggests that differences in the biofilm-forming abilities between type 3 fimbriae-expressing isolates is a consequence of variation in fimbriae regulation/expression mechanisms and/or the involvement of other biofilm factors, such as the capsule polysaccharide.
Intracellular levels of c-di-GMP coordinate type 3 fimbriae extension in K. pneumoniae
The best-studied usher-assembled pili/fimbriae are represented by the P (pap) pili and type 1 (fim) fimbriae, both of which are controlled by phase variation mechanisms ensuring their expression is either switched “ON” or “OFF” [13]. In the pap system, local and global DNA binding regulators direct phase switching through modulating DNA methylation patterns [31]. Phase variable expression of type 1 fimbriae in E. coli is coordinated by the inversion of a fimS DNA element located upstream of the fim operon, via site-specific recombinase-like integrases (FimB and FimE) [32], [33].
Quite unlike previously studied systems, the type 3 fimbriae of K. pneumoniae are regulated by c-di-GMP. A recent study identified MrkJ as a phosphodiesterase and demonstrated the activity of the purified protein in hydrolyzing c-di-GMP [34], and we demonstrated by HPLC analysis that MrkJ exhibits very strong phosphodiesterase activity. C-di-GMP was first identified as an allosteric regulator of cellulose synthase in Gluconacetobacter xylinus [35], [36], [37]. Since then, c-di-GMP has been characterized as an intracellular second messenger, ubiquitous in prokaryotes [38], [39], [40]. This important signaling molecule regulates a variety of processes, including motility, extracellular polysaccharide synthesis, cell differentiation and virulence gene expression [41], [42]. Deletion of mrkJ resulted in an increase in type 3 fimbriae production and biofilm formation as a result of the accumulation of intracellular c-di-GMP [34]. Furthermore, our transposon library screen has provided a mechanistic basis for understanding how c-di-GMP signaling controls biofilm formation and type 3 fimbriae in this emergent pathogen, given the opposing phenotypes of mutants defective in the GGDEF domain-containing diguanylate cyclase (YfiN) and EAL domain-containing phosphodiesterase (MrkJ), and the role of c-di-GMP in regulating the transcriptional activation mediated by the novel DNA-binding protein MrkH. Apart from MrkH, all three sequenced K. pneumoniae genomes contain only one other PilZ-domain protein – BcsA, the catalytic subunit of cellulose synthase. In addition, like most bacteria, K. pneumoniae harbours multiple GGDEF and EAL proteins (12 GGDEF, 9 EAL and 6 GGDEF-EAL). Rather than implicated in redundancy, these protein domains are usually coupled to sensory input or information transfer domains, and are thus believed to carry out specific tasks in the cell.
Some of the elements we have identified in the c-di-GMP-mediated ‘biofilm switch’ in K. pneumoniae have been reported in other bacteria, suggesting that at a cellular level, such a switch may represent a general control mechanism. For example, the Pseudomonas aeruginosa Cup (cup) fimbriae, assembled through the chaperone-usher pathway, are down-regulated at the transcriptional level indirectly by phosphodiesterase response regulators [43]. Furthermore, in E. coli, type 1 fimbriae expression in the adherent-invasive strain LF82 is also regulated, at least in part, via a c-di-GMP-dependent pathway [44].
The environmental signals that regulate YfiRNB expression to modulate type 3 fimbriae expression and hence biofilm formation, are unknown. In P. aeruginosa, it has been suggested that YfiB, which belongs to the Pal (peptidoglycan-associated lipoprotein) family of lipoproteins, could relay stress response signals from the outer envelope to YfiN via YfiR [45]. It is also understood that the periplasmic tyrosine phosphatase (TpbA), which responds to quorum sensing signals, can dephosphorylate a tyrosine residue located within the periplasmic domain of YfiN (termed TpbB) leading to deactivation of the GGDEF domain and c-di-GMP repression [46]. Whether analogous systems exist for the YfiRNB homolog in K. pneumoniae to control c-di-GMP levels, biofilm formation and type 3 fimbriae expression remains to be determined.
MrkH is predicted to contain a PilZ domain, widely distributed in bacterial proteins and responsible for c-di-GMP binding, which controls a range of c-di-GMP-mediated cellular functions [38], [47]. Several lines of evidence suggest that MrkH is a c-di-GMP effector protein. Firstly, the C-terminal PilZ domain of MrkH has all of the conserved residues known to be crucial for c-di-GMP binding. Secondly, changing the conversed arginine residue at position 113 within the putative c-di-GMP-binding domain of MrkH completely destroyed the ability of MrkH to activate the transcription from the mrkA promoter in vivo. Thirdly, the role of MrkH is in agreement with the phenotypes observed from mutational and over-expression experiments conducted with the c-di-GMP turnover enzymes YfiN and MrkJ. Fourthly, DNA-binding by purified MrkH is strongly stimulated in the presence of c-di-GMP.
MrkH is a novel transcriptional activator of the mrk gene cluster in K. pneumoniae
Transcriptional analysis using promoter-reporter fusions and a primer extension assay identified a single σ70 promoter (the mrkA promoter) preceding the mrkABCDF operon. Although the putative -10 region (TATATT) of this promoter matches well with the consensus sequence, the putative -35 region (TTAATG) and the spacer between the two regions (15 bp) are poorly conserved. The -35 region of σ70 promoters is recognized and contacted by region 4 of the RNA polymerase σ70 subunit during the first step of transcription initiation to form a closed complex [48]. With respect to the mrkA promoter, the presence of the poorly conserved -35 region and the shorter spacer (15 bp), which causes the imperfect alignment of the -35 and -10 regions, is the most likely explanation for the weak interaction of RNA polymerase with the promoter DNA and the resultant low basal levels of promoter activity that were observed in a MrkH– background. Remarkably, this weak promoter can be induced up to several hundred fold in bacterial strains expressing the MrkH protein, through the binding of the activator to a cis-acting element immediately upstream of the -35 region. The MrkH binding-site resembles the Class 1 Crp-dependent promoters of E. coli, where Crp activates transcription initiation by recruiting RNA polymerase to the promoter via a direct contact between Crp and the σ subunit of RNA polymerase [49].
MrkH is activated in the presence of c-di-GMP to ‘switch on’ biofilm formation
The PilZ domain characteristically undergoes significant structural changes upon c-di-GMP binding, allosterically activating the protein to interact with downstream effectors [50], [51], [52], [53]. We suggest that c-di-GMP binding to the PilZ domain of MrkH causes structural changes to the protein and activation of an output domain, specifically to permit DNA-binding of MrkH immediately upstream of the -35 box of the mrkA promoter. This complex could then stabilize or recruit RNA polymerase to the promoter for transcriptional initiation of the mrkABCDF operon, thereby effectively ‘switching on’ type 3 fimbriae production and biofilm formation. MrkH could represent a novel type of DNA-binding protein: it has no obvious homology to a HTH DNA-binding domain, which is most commonly used for DNA-binding by bacterial regulatory proteins. However, the N-terminal portion of the MrkH protein (the first 106 aa) is predicted by secondary structure prediction programs (such as PSIPRED [54]) to constitute five β strands flanked by two α helices which is similar to that of a bacterial LytTR DNA-binding domain [55], [56]. LytTR-domain proteins account for approximately 3% of all prokaryotic response regulators [57] and members include the Staphylococcus aureus AgrA, a global regulator of virulence [58], [59]; P. aeruginosa AlgR, which modulates the production of alginate [60]; and, interestingly, K. pneumoniae MrkE, a putative regulatory protein encoded on a plasmid next to the mrkABCDF gene cluster from isolate IA565 [9]. It is also suggested that a LytTR domain is structurally homologous to the well-characterized DNA-binding domain of the Sac7d protein from Sulfolobus acidocaldarius, which is comprised of five β strands followed by an α helix [55], [61], [62]. Although there is a lack of sequence similarity between various characterized LytTR DNA-binding motifs, the LytTR domain appears to share a common secondary structure which gives rise to a unique fold. We are currently investigating the structural and functional basis of any possible DNA-binding motif of MrkH.
C-di-GMP participates in the transcriptional regulation of other regulatory proteins. The transcriptional regulator VpsT from V. cholerae is a master regulator of biofilm formation that binds c-di-GMP (via a conserved W[F/L/M][T/S]R sequence motif) to inversely control extracellular matrix production and motility [63]. In the plant pathogen, Xanthomonas axonopodis, c-di-GMP allosterically represses the activity of Clp, a global transcriptional regulator of about 300 genes involved in pathogenesis [64], [65]. C-di-GMP association with the cyclic nucleotide monophosphate (cNMP) binding domain of Clp results in conformational changes that abolish the binding of Clp to its DNA targets. In P. aeruginosa, c-di-GMP binding to FleQ, a transcriptional repressor of the pel operon involved in exopolysaccharide substance production, inactivates its DNA binding ability to upregulate EPS biosynthesis [66], [67].
Although c-di-GMP has been implicated in numerous studies to mediate biofilm formation, this is the first demonstration that this compound can work as an effector that stimulates the DNA-binding ability of a PilZ domain-containing transcriptional activator. Therefore, our study identified the MrkH regulator as a missing link between the second messenger c-di-GMP and the formation of biofilms in K. pneumoniae. The dependence of type 3 fimbrial synthesis on the MrkH transcriptional activator, with its activity directly controlled by c-di-GMP via the expression of diguanylate cyclase and phosphodiesterase enzymes, permits an elegantly and tightly controlled mechanism by which K. pneumoniae can quickly regulate the switch between the planktonic and biofilm lifestyles when the relevant environmental sensors are triggered.
Close homologs of the mrkHIJ-ABCDF cluster are present in the C. koseri BAA-895 genome. Examination of the C. koseri MrkH, MrkI and MrkJ homologs revealed conservation of the PilZ, LuxR and EAL domains, respectively (data not shown). A close homolog of YfiN carrying a conserved GGDEF domain was also found (data not shown). C. koseri, a Gram-negative enterobacterium, is a cause of urinary tract infections, sepsis and meningitis, predominately amongst neonates and immunocompromised adults [68]. The habitat and epidemiology of this pathogen closely parallels that of K. pneumoniae, and expression of type 3 fimbriae to facilitate biofilm formation has been demonstrated [24], [69]. Given these similarities between the two pathogens, we speculate that the regulation of type 3 fimbriae expression in C. koseri might involve analogous mechanisms to those described here for K. pneumoniae.
MrkI is a candidate regulator of K. pneumoniae type 3 fimbriae function and expression
The MrkI regulator belongs to a family of transcriptional regulators (FixJ/LuxR/UhpA) that contain a C-terminal HTH DNA-binding motif and a receiver domain at the N-terminal region [70]. CsgD, the analogous protein found in Salmonella enterica serovar Typhimurium and E. coli, is a master regulator of the multicellular (rdar) morphotype of S. typhimurium [71] and acts by positively regulating two extracellular matrix components: curli fimbriae, via activation of the csgBAC operon [72], [73] and cellulose, via activation of the DGC-encoding gene adrA [74], [75]. The VspT regulator of V. cholerae described above is a member of the FixJ/LuxR/CsgD family of response regulators and consists of an N-terminal receiver domain and C-terminal HTH DNA-binding domain [63]. The c-di-GMP binding motif of VspT was found to be absent in MrkI, however.
We propose that MrkI, which is co-coordinately expressed with MrkH, possesses multiple functions to both regulate the expression and functionality of type 3 fimbriae. As a minor activator of type 3 fimbriae expression, at least three mechanisms are possible, including self-transcriptional activation with mrkH to increase c-di-GMP effector protein production, activation of DGC-encoding gene(s) to stimulate c-di-GMP production, or regulation of the fimbriae themselves through partial activation of the mrkABCDF operon. Thus, while we have demonstrated that MrkI is necessary for expression of functional type 3 fimbriae, further studies are required to determine which gene(s) are directly regulated by MrkI to eventuate in increased expression of type 3 fimbriae.
Implications and future research
The genetic screen reported here revealed the significance of c-di-GMP-mediated regulation of biofilm formation and type 3 fimbriae expression in K. pneumoniae (Figure 13). Type 3 fimbriae are important for efficient surface attachment and biofilm formation and are likely to be a primary virulence factor, with biofilms implicated as a significant cause of hospital-acquired infections and related deaths [76], [77]. Indwelling medical devices, such as urinary catheters, provide an attractive surface for bacterial pathogens such as K. pneumoniae to establish biofilms for colonization within human hosts. Future research lies not only in understanding how c-di-GMP effector proteins relay upstream signals to control phenotypes involved in biofilm formation, but in identifying the environmental signals that trigger c-di-GMP synthesis and breakdown. Moreover, c-di-GMP regulation and recognition within bacteria offer new targets for strategic intervention, for example the development of novel inhibitors that could be incorporated into the materials used to produce hospital devices.
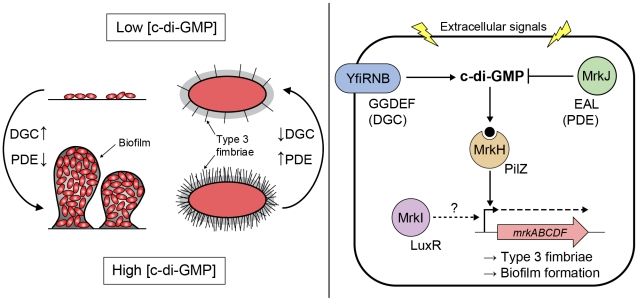
Figure 13. Model of c-di-GMP-mediated control of type 3 fimbriae expression and biofilm formation in K. pneumoniae.
Signals resulting in increased/decreased intracellular concentration of c-di-GMP, via changes in the relative activities of DGCs (carrying a GGDEF domain [YfiN]) and PDEs (carrying an EAL domain [MrkJ]), direct the DNA-binding activity of the c-di-GMP receptor MrkH (carrying a PilZ domain). High c-di-GMP levels promote biofilm formation through MrkH:c-di-GMP-dependent transcriptional activation of the mrkABCDF operon encoding type 3 fimbriae. The major pilin subunit MrkA is bound by the MrkB chaperone in the periplasm to activate it for polymerization by the usher translocase MrkC. Once sufficiently elongated, the fimbriae would emerge through the extracellular capsule layer (represented by grey shading surrounding the cell). Conversely, low c-di-GMP levels promote biofilm dispersal and the planktonic state through a decrease in activated MrkH:c-di-GMP availability. Whether the LuxR-like regulator MrkI acts on mrkABCDF gene expression, or the expression of other factors influencing type 3 fimbriae polymerization or capsule polysaccharide production remains to be determined.
Materials and Methods
Bacterial strains, plasmids and growth conditions
The bacterial strains and plasmids used in this study are described in Table 2. K. pneumoniae strain AJ218 (capsule serotype K54) is a human, urinary tract infection isolate [20]. K. pneumoniae AJ218Rif is a spontaneous rifampacin resistant strain used in conjugation experiments to create the transposon mutant library. E. coli DH5α was used for cloning purposes [78]. Unless otherwise stated, bacteria were maintained in Luria-Bertani (LB) medium overnight at 37°C with shaking. When appropriate, media were supplemented with antibiotics at the following concentrations: ampicillin (Ap), 100 µg/mL; kanamycin (Km), 50 µg/mL; chloramphenicol (Chl), 30 µg/mL (for E. coli DH5α) and 80 µg/mL (for K. pneumoniae); rifampacin (Rif), 150 µg/mL; trimethoprim (Tm), 40 µg/mL.
Table 2. Bacterial strains and plasmids used in this study.
| Strain or plasmid | Relevant phenotypes and genotypes | Source or reference |
| K. pneumoniae | ||
| AJ218 | Wild-type, clinical isolate, serotype K54; ApR | [20] |
| AJ218Rif | AJ218; RifR ApR | This study |
| JW30A8 | AJ218Rif mini-Tn5 mutant mrkA::Tn5Km2; ApR KmR | This study |
| JW69A4 | AJ218Rif mini-Tn5 mutant mrkB::Tn5Km2; ApR KmR | This study |
| JW8E7 | AJ218Rif mini-Tn5 mutant mrkC::Tn5Km2; ApR KmR | This study |
| JW34H5 | AJ218Rif mini-Tn5 mutant mrkH::Tn5Km2; ApR KmR | This study |
| JW64B4 | AJ218Rif mini-Tn5 mutant mrkI::Tn5Km2; ApR KmR | This study |
| JW45H9 | AJ218Rif mini-Tn5 mutant mrkJ::Tn5Km2; ApR KmR | This study |
| JW34D8 | AJ218Rif mini-Tn5 mutant yfiN::Tn5Km2; ApR KmR | This study |
| JW100 | AJ218 deletion mutant ΔmrkA::km; ApR KmR | This study |
| JW133 | AJ218 deletion mutant ΔmrkH; ApR | This study |
| JW96 | AJ218 deletion mutant ΔmrkI::km; ApR KmR | This study |
| JW99 | AJ218 deletion mutant ΔmrkJ::km; ApR KmR | This study |
| JW98 | AJ218 deletion mutant ΔyfiRNB::km; ApR KmR | This study |
| E. coli | ||
| S17 λpir | recA thi pro hsdR M + RP4::2-Tc::Mu::Km Tn7 λpir lysogen; TpR SmR | [95] |
| DH5α | F- endA1 hsdR17 (rk −, mk +) supE44 thi-1 λ− recA1 gyrA96 relA1 deoR Δ(lacZYA-argF)-U169 Φ80dlacZΔM15; NalR | [78] |
| MC4100 | Δ(argF-lac)U169, rpsL150, relA, araD139, fib5301, deoC1, ptsF25 | [96] |
| BL21(DE3) | F-, ompT, hsdSB(rB-, mB-), dcm, gal, λ(DE3) | [97] |
| Plasmids | ||
| pMrk | AJ218 mrkABCDF cloned into pACYC184; ChlR | This study |
| pMrkH | AJ218 mrkH cloned into pACYC184; ChlR | This study |
| pMrkI | AJ218 mrkI cloned into pACYC184; ChlR | This study |
| pMrkJ | AJ218 mrkJ cloned into pACYC184; ChlR | This study |
| pYfiRNB | AJ218 yfiRNB cloned into pACYC184; ChlR | This study |
| pACYC184 | Medium-copy-no. cloning vector, p15A ori; TetR ChlR | [98] |
| pBR322 | Medium-copy-no. cloning vector, pMB1 ori; TetR ApR | [99] |
| pGEM-T Easy | High-copy-no. cloning vector for PCR products; ApR | Promega |
| TOPO-TA | High-copy-no. cloning vector for PCR products; ApR KmR | Invitrogen |
| pUT-miniTn5Km2 | Mini-Tn5Km2 transposon in the pUT vector; ApR KmR | [100] |
| pKD4 | Source of FRT-flanked KanR cassette; ApR KmR | [80] |
| pACBSR | Ara promoter control, I-SceI and λ Red recombinase; ChlR | [79] |
| pCP20 | Flp helper plasmid, temp-sensitive replication; ApR ChlR | [81] |
| pMU2385 | galK'-lac'Z, IncW, single-copy-no. transcriptional-fusion vector; TpR | [101] |
| pKK232.8 | pBR322 derivative used for making promoter-cat fusions; ApR ChlR | Amersham |
| mrkA-lacZ-1 | mrkA-lacZ transcriptional fusion vector (pMU2385) from mrkA promoter nucleotides −759 to +109 relative to the mrkA start codon; TpR | This study |
| mrkA-lacZ-2 | mrkA-lacZ transcriptional fusion vector (pMU2385) from mrkA promoter nucleotides −295 to +109 relative to the mrkA start codon; TpR | This study |
| mrkA-lacZ-3 | mrkA-lacZ transcriptional fusion vector (pMU2385) from mrkA promoter nucleotides −295 to −165 relative to the mrkA start codon; TpR | This study |
| mrkA-lacZ-4 | mrkA-lacZ transcriptional fusion vector (pMU2385) from mrkA promoter nucleotides −259 to +109 relative to the mrkA start codon; TpR | This study |
| mrkA-cat | mrkA-cat transcriptional fusion vector (pKK232.8) from mrkA promoter nucleotides −295 to +109 relative to the mrkA start codon; ApR ChlR | This study |
| pET11a | Expression vector using T7 promoter; ApR | Novagen |
| pET11a-mrkH-8His | AJ218 mrkH tagged with 8×His at C-terminus cloned into pET11a; ApR | This study |
| pET11a-mrkH(113R-A)-8His | AJ218 mrkH:113R-A tagged with 8×His at C-terminus cloned into pET11a; ApR | This study |
| pET11a-mrkJ-8His | AJ218 mrkJ tagged with 8×His at C-terminus cloned into pET11a; ApR | This study |
| pGMrkH-8His | AJ218 mrkH tagged with 8×His at C-terminus cloned into pGEM-T Easy; ApR | This study |
| pGMrkH(113-R)-8His | AJ218 mrkH carrying R→A amino acid substitution at residue position 113 tagged with 8×His at C-terminus cloned into pGEM-T Easy; ApR | This study |
| pMrkH(113R-A) | AJ218 mrkH carrying R→A amino acid substitution at residue position 113 cloned into pACYC184; ChlR | This study |
| pMrkJ(36ECL-AAA) | AJ218 mrkJ carrying ECL→AAA amino acid substitution starting at residue position 36 cloned into pACYC184; ChlR | This study |
| pYfiRNB(328DEF-AAA) | AJ218 yfiRNB carrying DEF→AAA amino acid substitution starting at residue position 328 of YfiN cloned into pACYC184; ChlR | This study |
| pBMrkJ(36ECL-AAA) | AJ218 mrkJ carrying ECL→AAA amino acid substitution starting at residue position 36 cloned into pBR322; ApR | This study |
| pBYfiRNB(328DEF-AAA) | AJ218 yfiRNB carrying DEF→AAA amino acid substitution starting at residue position 328 of YfiN cloned into pBR322; ApR | This study |
DNA manipulation techniques
PCR amplifications were performed using GoTaq Green Master Mix (Promega, Madison, WI), Phusion Flash High-Fidelity PCR Master Mix (Finnzymes, Finland) or Vent DNA Polymerase (New England Biolabs, Ipswich, MA). Restriction endonucleases and T4 DNA ligase were obtained from New England Biolabs. Synthetic oligonucleotides for PCR and sequencing (Table S1) were obtained from GeneWorks (Hindmarsh, South Australia, Australia).
Transposon mutagenesis and Y-linker ligation PCR
Mid-log cultures of donor (E. coli S17-λpir harboring suicide vector pUT-mini-Tn5Km2) and recipient (K. pneumoniae AJ218Rif) were mixed in a 1∶1 ratio in a final volume of 1 mL. The conjugation mix was then centrifuged, resuspended in 0.1 mL LB and grown on LB agar for 6 h. Bacterial lawn growth was resuspended in LB, diluted 1∶100 and re-plated onto LB agar containing kanamycin and rifampacin. A total of 7,000 kanamycin and rifampacin resistant colonies were streak-diluted on LB agar containing kanamycin and subsequently stored in LB containing 10% glycerol and kanamycin at −70°C until required. The sequences flanking the transposon insertions were amplified by Y-linker ligation PCR [22].
Construction of K. pneumoniae deletion mutants
Knockout mutations, in which target genes were deleted by allelic exchange with a kanamycin resistance-encoding gene (km), were constructed in K. pneumoniae AJ218 using a “gene gorging” technique [79]. All primers used are listed in Table S1. ‘Donor’ plasmids carrying the desired mutation were constructed as follows. The km gene was amplified from pKD4 [80] using primers kanF and kanR. The resulting product included flanking fragment length polymorphism (FLP) recombinase target (FRT) sites to permit subsequent km excision [81]. Using K. pneumoniae AJ218 genomic DNA as the template, approximately 0.5 kb regions flanking the upstream and downstream sequence of the target gene were PCR amplified. The three fragments were joined together, with the km gene flanked by the upstream and downstream target gene sequences, in equimolar amounts using overlapping extension PCR [82]. The ISce-I-flanked PCR products were cloned into pGEM-T Easy (Promega) to yield donor plasmids, which were then sequenced.
The ‘mutagenesis’ plasmid pACBSR carries genes encoding I-SceI endonuclease and lambda Red recombinase under inducible control by L-arabinose [79]. Donor plasmids and pACBSR were transformed into electrocompetent K. pneumoniae AJ218 cells (0.1 cm gap-width cuvette; 200 ohms, 25 µF, 1.8 kV) and selected on LB agar containing kanamycin and chloramphenicol. A single co-transformant was inoculated into 1 mL LB containing 0.2% L-arabinose (Sigma-Aldrich, St. Louis, MO) and chloramphenicol and grown in a shaking incubator at 37°C for 16 h. Cell dilutions were grown on LB agar containing kanamycin, and resultant colonies were screened by colony PCR using primers flanking the targeted region and within the km gene. The loss of pACBSR was induced by 0.2% L-arabinose without selection. When required, the km gene was excised via the FRT sites using the FLP helper plasmid pCP20 [81].
Creation of complementation constructs
Wild-type K. pneumoniae AJ218 genes were PCR amplified (Table S1), cloned into pGEM-T Easy and sequenced, followed by restriction enzyme digestion and insertion within the tetracycline resistance-encoding gene (tet) of pACYC184 via unique BamHI/SalI restriction sites. Apart from pMrkI, in which mrkI was transcribed from the tet gene promoter of pACYC184, all complement plasmids contained AJ218 genes that were transcribed from their native promoter, in the opposite orientation of the tet gene. Constructs were maintained in cells with chloramphenicol resistance selection.
Construction of site-directed mutant constructs
Site-directed mutant alleles of mrkH, mrkJ and yfiN were constructed by overlapping-extension PCR [82] of wild-type K. pneumoniae AJ218 genomic DNA template using mutagenic oligonucleotides (Table S1). Overlapping primers were used together with the relevant upstream or downstream complementation primer. Amplified fragments were cloned into pGEM-T Easy and sequenced, followed by restriction enzyme digestion and insertion within the tet gene of pACYC184 or pBR322 via unique BamHI/SalI restriction sites.
Static biofilm assays
Biofilm assays were performed according to O'Toole with minor modifications [21]. Transposon mutants were incubated statically overnight at room temperature in 96-well plates containing LB and kanamycin and subsequently diluted 1∶50 in 100 µL M63B1-GCAA minimal media (containing 1% glycerol and 0.3% casamino acids) in duplicate 96-well, flat bottom, non-tissue culture treated, polyvinyl chloride microtiter plates (Falcon; BD Biosciences, San Jose, CA). Wells containing growth medium alone were used as negative controls and the measurements of these wells were subtracted from the experimental measurements. Following 8 h static incubation at 37°C, planktonic bacteria were decanted and wells were washed twice with distilled water. Biofilms attached to well surfaces were stained for 15 min at room temperature with 125 µL of 0.1% (wt/vol) crystal violet solution (Sigma-Aldrich). The crystal violet solution was decanted and wells were subsequently washed twice with distilled water to remove traces of unbound dye. The bound dye was solubilized from adherent cells with 33% acetic acid and subsequently quantified by measuring the optical density at 595 nm. This assay was similarly applied to examine the formation of biofilms by gene deletion mutants, however strains were initially grown in 10 mL LB overnight in shaking conditions at 37°C and the length of the biofilm assay was 24 h before biofilms were stained and quantified. The data for each strain represent average values taken from four replicate wells performed in two independent experiments.
Flow cell biofilm assays
For flow cell assays, biofilm culture media was a 1∶2,000 dilution of M63B1-GCAA culture media used for static biofilm assays. Biofilms were cultivated at 37°C in three-channel flow cells with individual channel dimensions of 1×4×40 mm (BioCentrum-DTU, Denmark). An ibidi cover slip (ibidi GmbH, Germany) was used as the substratum for biofilm growth. Flow cells were inoculated with overnight LB cultures diluted 1∶100 in warm culture media containing antibiotics as appropriate for plasmid-containing strains, and allowed to attach for 1 h at 37°C without flow. Flow was then commenced and maintained at 151 µL/min for 96 h. Biofilms were stained with 1.25 mM Syto64 (Invitrogen, Carlsbad, CA) to visualize cells and fixed with 4% paraformaldehyde.
Microscopy and image analysis
Microscopic observations of biofilms and image acquisitions were performed utilizing confocal scanning laser microscopy (CSLM) on a Nikon A1 confocal microscope, using a Plan Apo 40×/1.42 oil objective. Images were reconstructed from Z-sections and rendered for 3D visualization using the IMARIS software package (Bitplane AG, Zurich, Switzerland). For statistical evaluation of biofilm structures, duplicate flow cells were prepared for each strain, and at least six image stacks (0.5 µm slices) per flow chamber were obtained for the mature biofilm at 96 h post-inoculation. The images were analyzed with the biofilm computer program COMSTAT [83].
Detection of fimbriae expression
The presence of type 3 fimbriae was determined by mannose resistant hemagglutination (MRHA) assays [8]. Human erythrocytes (group A) were tanned by incubating equal volumes of 0.1% (wt/vol) tannic acid (Sigma-Aldrich) solution in saline and a 3% erythrocyte suspension in PBS for 15 min at 37°C. The erythrocytes were subsequently washed twice in PBS. Bacteria were grown overnight in shaking conditions in LB and subsequently washed twice and resuspended in PBS to approximately 1×1010 CFU/mL. A series of 2-fold dilutions of the bacterial suspension with or without 4% D-mannose (Sigma-Aldrich) was mixed with equal volumes (25 µL) of tanned erythrocytes in the depressions of porcelain tiles. The plates were rocked gently for 10 min at room temperature, after which the minimum bacterial density (CFU/mL) required to agglutinate erythrocytes was measured.
Type 3 fimbriae expression was also detected by immunoblot analysis using polyclonal rabbit antiserum prepared against purified MrkA. Whole cell lysates were prepared from 48 h M63B1-GCAA (containing 1% glycerol and 0.3% casamino acids) static cultures. Samples were separated by sodium dodecyl sulphate (SDS)-polyacrylamide gel electrophoresis (PAGE) and transferred to Hybond-C Extra nitrocellulose (Amersham Biosciences, Sweden) using a Trans-Blot SD Electrophoretic Transfer Cell (Bio-Rad Laboratories, Hercules, CA) at 12 V for 30 min. Goat anti-rabbit IgG-HRP (Invitrogen) was used as the secondary antibody at a concentration of 1∶3,000. Membranes were developed with TMB Membrane Peroxidase Substrate (KPL, Gaithersburg, MD). Loading was normalized by quantifying and comparing the relative differences of samples first separated by SDS-PAGE and stained with Coomassie-blue. The stained gel was scanned using a Kodak Digital Imaging System 4000MM (Eastman Kodak Company, Hemel Hempstead, England) and the intensity of a strongly expressed house-keeping gene product was measured to normalize the loading of samples in the subsequent immunoblot experiment.
RNA preparation and RT-PCR conditions
Total RNA was extracted from K. pneumoniae AJ218 (grown to mid-log phase, OD600 = 0.8) using a FastRNA Pro Blue Kit (Qbiogene, Irvine, CA) and residual DNA was eliminated with a TURBO DNA-free Kit (Applied Biosystems/Ambion, Carlsbad, CA) according to the manufacturer's recommendations. First-strand cDNA synthesis was performed using 200 U SuperScript II Reverse Transcriptase (Invitrogen) using 3 µg total RNA and 20 pmole of the reverse primer: mrkI-R or mrkJ-R (Table S1). PCR of cDNA templates was performed using GoTaq Green Master Mix. Removal of contaminating DNA from the RNA sample was verified by PCR in the absence of reverse transcription. Reactions were separated by 1% (wt/vol) agarose gel electrophoresis.
Quantitative RT-PCR
RNA was extracted from K. pneumoniae AJ218 strains and residual DNA eliminated, as stated above. cDNA was synthesized using 70 ng of random hexamers (Invitrogen) and Superscript II Reverse Transcription kit (Invitrogen), according to the manufacturer's guidelines. qPCR was performed using SYBR green (Quantace; SensiMix) and the primer pairs mrkA127F and mrkA265R and rpoD562F and rpoD677R (used at 0.125 µM each; Table S1). Cycling conditions were as follows: 95°C for 10 min followed by 35 cycles of 95°C for 30 sec, 55°C for 60 sec and 72°C for 15 sec. Primer efficiencies (E = 10(−1/slope) under these conditions were calculated following real-time PCR on 10-fold serially diluted genomic DNA, where the co-efficient for the standard curve was greater than 0.98. The efficiency-corrected, relative gene expression was determined using the Stratagene Mx3005 qPCR Thermocycler (Agilent Technologies, La Jolla, CA) whereby the expression of mrkA was normalised to the expression of rpoD. Primer specificity was determined by melting curve analysis and no Ct value was recorded for no template or no reverse transcriptase controls. All qPCR sample reactions were performed in triplicate.
Construction of lacZ and cat transcriptional fusions
The lacZ and cat transcriptional fusions were constructed by PCR amplification of desirable DNA fragments using chromosomal DNA of K. pneumoniae AJ218 as template and the primers described in Table S1. PCR fragments were cloned into TOPO-TA (Invitrogen) or pGEM-T Easy cloning vectors and sequenced. The fragments were then each excised from the TOPO or pGEM derivatives and cloned into appropriate sites within plasmids pMU2385 and pKK232.8, to create lacZ and cat transcriptional fusions.
Primer extension
Total cellular RNA was extracted (as stated above) from E. coli MC4100 strains containing pMrkH with either pMU2385 (control) or mrkA-lacZ-2. Cells were grown to mid-log phase (OD600 = 0.6). Primer Px1mrkARev (Table S1) was labeled at the 5′ end with both [γ-32P]ATP (Perkin Elmer, Waltham, MA) and T4 polynucleotide kinase (Promega) and subsequently co-precipitated with 5 µg of total RNA isolated from E. coli strain MC4100 containing pMrkH with either pMU2385 (control) or mrkA-lacZ-2. Hybridization was carried out at 45°C for 15 min in 10 mL of TE buffer containing 150 mM KCl. Primer extension reactions were started by the addition of 24 µL of extension solution (20 mM Tris HCl [pH 8.4], 10 mM MgCl2, 10 mM DTT, 2 mM dNTPs and 1 U/mL AMV Reverse Transcriptase) and were carried out at 42°C for 60 min. Samples were then precipitated and analyzed on a sequencing gel.
β-galactosidase and CAT assays
β-galactosidase activity was assayed as described elsewhere [84]. Specific activity was expressed in units described therein. The data are the results of at least three independent assays. The CAT activity of mid-log-phase cultures, grown in LB, was assayed as described elsewhere [85]. The cells were disrupted by sonication, and cellular debris was removed by centrifugation before the assays were carried out. Each assay was performed at least three times. CAT activity was expressed as units per milligram of protein.
Expression and purification of MrkH-8×His, MrkH(113R-A)-8×His and MrkJ-8×His
The coding regions of mrkH, mrkH:113R-A and mrkJ flanked by NdeI and BamHI sites were PCR amplified using primer pairs mrkH(NdeI)11a and mrkH(BamHI)11a for mrkH and mrkH:113R-A, and mrkJ(NdeI)11a and mrkJ(BamHI)11a for mrkJ (Table S1) using pMrkH, pMrkH(113R-A) or pMrkJ as template, respectively. The amplified DNA fragments were cloned into TOPO-TA and sequenced. The mrkH, mrkH:113R-A and mrkJ fragments encoding the MrkH, MrkH:113R-A and MrkJ proteins with eight histidine residues tagged at the C-terminal end were then excised and cloned into the NdeI and BamHI sites of pET11a (Novagen, Madison, WI) to form pET11a-mrkH-8His, pET11a-mrkH(113R-A)-8His and pET11a-mrkJ-8His. For over-expression of His-tagged proteins, E. coli expression strain BL21(DE3) containing pET11a-constructs was induced with 0.3 mM isopropyl-β-D-thiogalactopyranoside (IPTG) for 3 h at 20°C (for MrkH-8×His and MrkH(113R-A)-8×His) or at 16°C (for MrkJ-8×His). Over-expressed proteins were purified using Metal Affinity Chromatography.
Electrophoretic mobility shift assay (EMSA)
Primer Px1mrkARev was labeled at the 5′ end with [γ-32P]ATP and T4 polynucleotide kinase. The DNA fragment containing the mrkA regulatory region was generated by PCR using 32P-labelled primers Px1mrkARev and mrk295F, with TOPO-TA carrying the mrkA regulatory region as template. Each end-labeled fragment was incubated with varying amounts of purified MrkH-8×His and MrkH(113R-A)-8×His protein with or without 200 µM c-di-GMP (enzymatically synthesized; for details see [86]) or varying amounts of purified MrkH-8×His with 200 µM GTP at 30°C for 20 min in the binding buffer (10 mM Tris HCl [pH 7.4], 50 mM KCl, 1 mM DTT, 100 µg/mL BSA and 5 ng/µL poly[dI-dC]). Glycerol was added to a final concentration of 6.5%. Samples were analyzed by electrophoresis on 5% native polyacrylamide gels (37.5∶1) containing 50 µM c-di-GMP. Electrophoresis was carried out at room temperature for approximately 8 h at 10 V/cm.
The binding affinities of PilZ-domain proteins with c-di-GMP are typically below 10 µM [50], [63], [87], [88]. To compensate for the loss of c-di-GMP through diffusion into the polyacrylamide gel and running buffer during electrophoresis, saturating concentrations of c-di-GMP (200 µM) were used to ensure that all MrkH molecules were ligand-bound. Saturating ligand concentrations have been used in other EMSA studies of protein interactions with c-di-GMP [63] and cAMP [89], [90].
High-Performance Liquid Chromatography (HPLC)
The procedure for measuring phosphodiesterase activity has been described previously [86], [91]. Briefly, the purified MrkJ-8×His was incubated with 100 µM of c-di-GMP in 100 mM Tris buffer (pH 8.0) with 50 mM KCl and 25 mM MgCl2 at room temperature. Reactions were stopped by heating the reaction mixture at 95°C for 10 min. After the protein precipitate was removed by centrifugation at 135,000 rpm for 10 min, the supernatant was analyzed by HPLC. The formation of 5′-pGpG was monitored using an Agilent LC1200 system equipped with an XDB C18 column (4.6×150 mm) (mobile phase: 20 mM triethylammonium bicarbonate [pH 7.0], 10% methanol, 1 mL/min).
Statistical analyses
All statistical analyses were performed using JMP 8.0 software (SAS Institute, Cary, NC). Data were log10/sqrt transformed for normality and analyzed by ANOVA with Tukey HSD post-hoc comparisons performed when necessary. Where normal distributions could not be met, non-parametric Van der Waerden tests were performed with Tukey HSD post-hoc comparisons when necessary. Where values were obtained as below the detection limit of an assay, the minimum detectable value minus 1 was assigned. P<0.05 was considered significant.
Accession numbers
K. pneumoniae AJ218 gene sequences were deposited in GenBank under the accession numbers JF759917 (mrkH), JF759918 (mrkI), JF759919 (mrkJ), JF759920 (yfiN), and JF759921 (mrkABCDF).
Supporting Information
Biofilm formation by K. pneumoniae AJ218. Biofilm formation by K. pneumoniae AJ218 wild-type and isogenic mutants strains +/- empty pACYC184 plasmids. Biofilm formation was determined using the static microtiter plate assay following incubation in M63B1-GCAA minimal media (supplemented with 1% glycerol and 0.3% casamino acids) for 24 h under static conditions. Results are expressed as a percentage of the biofilm produced by the wild-type AJ218 strain, which is set to 100%. All values represent the mean of four replicate sample wells for each strain performed in two independent experiments. The error bars represent the standard deviation.
(TIF)
Type 3 fimbriae expression by K. pneumoniae AJ218. Mannose resistant Klebsiella-like hemagglutination (MR/K HA) by K. pneumoniae AJ218 wild-type and isogenic mutant strains +/- empty pACYC184 plasmids using human erythrocytes. MR/K HA titer is expressed as the lowest concentration (CFU/mL) of bacteria causing a visible agglutination reaction. Values represent the mean of three independent experiments. The error bars represent the standard deviation.
(TIF)
Coomassie-blue stained SDS-PAGE of over-expressed and purified MrkH-8×His (10 µg loaded). The recombinant MrkH-8×His protein (used for EMSA studies) is labeled, which migrates at approximately 28 kDa.
(TIF)
EMSA of the mrkA fragment. The buffers and conditions used in the assay are as described in the Materials and Methods. The 32P-labelled PCR fragment containing the mrkA regulatory region was generated using primer pairs 32P-Px1mrkARev and mrk295F. The mrkA fragment was mixed with varying amounts of either the purified wild-type MrkH-8×His protein (from 0 to 500 nM) in the presence of 200 μM of GTP (left panel) or the purified mutant MrkH(113R-A)-8×His protein (from 0 to 500 nM) in the presence of 200 μM of c-di-GMP (right panel). Following incubation at 30°C for 20 min, the samples were analyzed on native polyacrylamide gels. The unbound DNA bands (F) are marked.
(TIF)
Immunoblot of MrkH-8×His expression. Samples were prepared by sonication followed by centrifugation and supernatant samples were separated by SDS-PAGE. Following transfer, the membrane was probed with anti-His antibody. Shown are E. coli MC4100 strains harboring pGMrkH-8His (wild-type) and pGMrkH(113R-A)-8His (mutant) preparations. E. coli MC4100 was used as the negative control. MrkH-8×His is labeled, which migrates at approximately 28 kDa.
(TIF)
Coomassie-blue stained SDS-PAGE of over-expressed and purified MrkJ-8×His (10 µg loaded). MrkJ-8×His protein (used for HPLC studies) is labeled, which migrates at approximately 29 kDa.
(TIF)
Oligonucleotide primers used in this study.
(DOC)
Acknowledgments
The authors would like to gratefully acknowledge the support of members from the Strugnell Laboratory. We sincerely thank A. Walduck and K. Holt for critical reading of the manuscript, C. Kinnear for performing the statistical analysis, T. Scott for providing human erythrocytes and D. Hocking and K. Azzopardi from the Robins-Browne Laboratory for their assistance and suggestions.
Footnotes
The authors have declared that no competing interests exist.
The authors gratefully acknowledge support from research grants from the Australian National Health and Medical Research Council and the Australian Research Council. JJW is supported by a Melbourne Research Scholarship. CBW is supported by a NHMRC Senior Research Fellowship. OLW is supported by a NHMRC R. D. Wright Development Award. TL is supported by an ARC Federation Fellowship. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Carpenter JL. Klebsiella pulmonary infections: occurrence at one medical center and review. Rev Infect Dis. 1990;12:672–682. doi: 10.1093/clinids/12.4.672. [DOI] [PubMed] [Google Scholar]
- 2.Langley JM, Hanakowski M, LeBlanc JC. Unique epidemiology of nosocomial urinary tract infection in children. Am J Infect Control. 2001;29:94–98. doi: 10.1067/mic.2001.111537. [DOI] [PubMed] [Google Scholar]
- 3.Marschall J, Fraser VJ, Doherty J, Warren DK. Between Community and Hospital: Healthcare Associated Gram Negative Bacteremia among Hospitalized Patients. Infect Control Hosp Epidemiol. 2009;30:1050–1056. doi: 10.1086/606165. [DOI] [PubMed] [Google Scholar]
- 4.Mathai D, Jones RN, Pfaller MA. Epidemiology and frequency of resistance among pathogens causing urinary tract infections in 1,510 hospitalized patients: a report from the SENTRY Antimicrobial Surveillance Program (North America). Diagn Microbiol Infect Dis. 2001;40:129–136. doi: 10.1016/s0732-8893(01)00254-1. [DOI] [PubMed] [Google Scholar]
- 5.Sligl W, Taylor G, Brindley PG. Five years of nosocomial Gram-negative bacteremia in a general intensive care unit: epidemiology, antimicrobial susceptibility patterns, and outcomes. Int J Infect Dis. 2006;10:320–325. doi: 10.1016/j.ijid.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 6.Sohn AH, Garrett DO, Sinkowitz-Cochran RL, Grohskopf LA, Levine GL, et al. Prevalence of nosocomial infections in neonatal intensive care unit patients: Results from the first national point-prevalence survey. J Pediatr. 2001;139:821–827. doi: 10.1067/mpd.2001.119442. [DOI] [PubMed] [Google Scholar]
- 7.Yu VL, Hansen DS, Ko WC, Sagnimeni A, Klugman KP, et al. Virulence characteristics of Klebsiella and clinical manifestations of K. pneumoniae bloodstream infections. Emerg Infect Dis. 2007;13:986–993. doi: 10.3201/eid1307.070187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duguid JP. Fimbriae and adhesive properties in Klebsiella strains. J Gen Microbiol. 1959;21:271–286. doi: 10.1099/00221287-21-1-271. [DOI] [PubMed] [Google Scholar]
- 9.Allen BL, Gerlach GF, Clegg S. Nucleotide sequence and functions of mrk determinants necessary for expression of type 3 fimbriae in Klebsiella pneumoniae. J Bacteriol. 1991;173:916–920. doi: 10.1128/jb.173.2.916-920.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hornick DB, Allen BL, Horn MA, Clegg S. Adherence to respiratory epithelia by recombinant Escherichia coli expressing Klebsiella pneumoniae type 3 fimbrial gene products. Infect Immun. 1992;60:1577–1588. doi: 10.1128/iai.60.4.1577-1588.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hornick DB, Thommandru J, Smits W, Clegg S. Adherence properties of an mrkD-negative mutant of Klebsiella pneumoniae. Infect Immun. 1995;63:2026–2032. doi: 10.1128/iai.63.5.2026-2032.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tarkkanen AM, Allen BL, Westerlund B, Holthofer H, Kuusela P, et al. Type V collagen as the target for type-3 fimbriae, enterobacterial adherence organelles. Mol Microbiol. 1990;4:1353–1361. doi: 10.1111/j.1365-2958.1990.tb00714.x. [DOI] [PubMed] [Google Scholar]
- 13.Waksman G, Hultgren SJ. Structural biology of the chaperone–usher pathway of pilus biogenesis. Nat Rev Microbiol. 2009;7:765–774. doi: 10.1038/nrmicro2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jagnow J, Clegg S. Klebsiella pneumoniae MrkD-mediated biofilm formation on extracellular matrix- and collagen-coated surfaces. Microbiology. 2003;149:2397–2405. doi: 10.1099/mic.0.26434-0. [DOI] [PubMed] [Google Scholar]
- 15.Langstraat J, Bohse M, Clegg S. Type 3 fimbrial shaft (MrkA) of Klebsiella pneumoniae, but not the fimbrial adhesin (MrkD), facilitates biofilm formation. Infect Immun. 2001;69:5805–5812. doi: 10.1128/IAI.69.9.5805-5812.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sebghati TA, Clegg S. Construction and characterization of mutations within the Klebsiella mrkD1P gene that affect binding to collagen type V. Infect Immun. 1999;67:1672–1676. doi: 10.1128/iai.67.4.1672-1676.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tarkkanen AM, Virkola R, Clegg S, Korhonen TK. Binding of the type 3 fimbriae of Klebsiella pneumoniae to human endothelial and urinary bladder cells. Infect Immun. 1997;65:1546–1549. doi: 10.1128/iai.65.4.1546-1549.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boddicker JD, Anderson RA, Jagnow J, Clegg S. Signature-tagged mutagenesis of Klebsiella pneumoniae to identify genes that influence biofilm formation on extracellular matrix material. Infect Immun. 2006;74:4590–4597. doi: 10.1128/IAI.00129-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schembri M, Blom J, Krogfelt K, Klemm P. Capsule and fimbria interaction in Klebsiella pneumoniae. Infect Immun. 2005;73:4626–4633. doi: 10.1128/IAI.73.8.4626-4633.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jenney AW, Clements A, Farn JL, Wijburg OL, McGlinchey A, et al. Seroepidemiology of Klebsiella pneumoniae in an Australian Tertiary Hospital and its implications for vaccine development. J Clin Microbiol. 2006;44:102–107. doi: 10.1128/JCM.44.1.102-107.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Merritt JH, Kadouri DE, O'Toole GA. Growing and analyzing static biofilms. Curr Protoc Microbiol Chapter 1: Unit 1B.1. 2005 doi: 10.1002/9780471729259.mc01b01s00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kwon YM, Ricke SC. Efficient amplification of multiple transposon-flanking sequences. J Microbiol Methods. 2000;41:195–199. doi: 10.1016/s0167-7012(00)00159-7. [DOI] [PubMed] [Google Scholar]
- 23.Marchler-Bauer A, Anderson J, Chitsaz F, Derbyshire M, DeWeese-Scott C, et al. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 2009;37:D205–D210. doi: 10.1093/nar/gkn845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ong CL, Beatson SA, Totsika M, Forestier C, McEwan AG, et al. Molecular analysis of type 3 fimbrial genes from Escherichia coli, Klebsiella and Citrobacter species. BMC Microbiol. 2010;10:183. doi: 10.1186/1471-2180-10-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller MB, Bassler BL. Quorum sensing in bacteria. Annu Rev Microbiol. 2001;55:165–199. doi: 10.1146/annurev.micro.55.1.165. [DOI] [PubMed] [Google Scholar]
- 26.Di Martino P, Cafferini N, Joly B, Darfeuille-Michaud A. Klebsiella pneumoniae type 3 pili facilitate adherence and biofilm formation on abiotic surfaces. Res Microbiol. 2003;154:9–16. doi: 10.1016/s0923-2508(02)00004-9. [DOI] [PubMed] [Google Scholar]
- 27.Nuccio SP, Bäumler AJ. Evolution of the chaperone/usher assembly pathway: fimbrial classification goes Greek. Microbiol Mol Biol Rev. 2007;71:551–575. doi: 10.1128/MMBR.00014-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Remaut H, Tang C, Henderson N, Pinkner J, Wang T, et al. Fiber formation across the bacterial outer membrane by the chaperone/usher pathway. Cell. 2008;133:640–652. doi: 10.1016/j.cell.2008.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Old DC, Adegbola RA. Antigenic relationships among type-3 fimbriae of Enterobacteriaceae revealed by immunoelectronmicroscopy. J Med Microbiol. 1985;20:113–121. doi: 10.1099/00222615-20-1-113. [DOI] [PubMed] [Google Scholar]
- 30.Old DC, Tavendale A, Senior BW. A comparative study of the type-3 fimbriae of Klebsiella species. J Med Microbiol. 1985;20:203–214. doi: 10.1099/00222615-20-2-203. [DOI] [PubMed] [Google Scholar]
- 31.van der Woude M, Braaten B, Low D. Epigenetic phase variation of the pap operon in Escherichia coli. Trends Microbiol. 1996;4:5–9. doi: 10.1016/0966-842x(96)81498-3. [DOI] [PubMed] [Google Scholar]
- 32.Gally DL, Leathart J, Blomfield IC. Interaction of FimB and FimE with the fim switch that controls the phase variation of type 1 fimbriae in Escherichia coli K-12. Mol Microbiol. 1996;21:725–738. doi: 10.1046/j.1365-2958.1996.311388.x. [DOI] [PubMed] [Google Scholar]
- 33.McClain MS, Blomfield IC, Eisenstein BI. Roles of fimB and fimE in site-specific DNA inversion associated with phase variation of type 1 fimbriae in Escherichia coli. J Bacteriol. 1991;173:5308–5314. doi: 10.1128/jb.173.17.5308-5314.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson JG, Clegg S. The role of MrkJ, a phosphodiesterase, in type 3 fimbrial expression and biofilm formation in Klebsiella pneumoniae. J Bacteriol. 2010;192:3944–3950. doi: 10.1128/JB.00304-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mayer R, Ross P, Weinhouse H, Amikam D, Volman G, et al. Polypeptide composition of bacterial cyclic diguanylic acid-dependent cellulose synthase and the occurrence of immunologically crossreacting proteins in higher plants. Proc Natl Acad Sci U S A. 1991;88:5472–5476. doi: 10.1073/pnas.88.12.5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ross P, Weinhouse H, Aloni Y, Michaeli D, Weinberger-Ohana P, et al. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature. 1987;325:279–281. doi: 10.1038/325279a0. [DOI] [PubMed] [Google Scholar]
- 37.Weinhouse H, Sapir S, Amikam D, Shilo Y, Volman G, et al. c-di-GMP-binding protein, a new factor regulating cellulose synthesis in Acetobacter xylinum. FEBS Lett. 1997;416:207–211. doi: 10.1016/s0014-5793(97)01202-7. [DOI] [PubMed] [Google Scholar]
- 38.Hengge R. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol. 2009;7:263–273. doi: 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- 39.Jenal U, Malone J. Mechanisms of cyclic-di-GMP signaling in bacteria. Annu Rev Genet. 2006;40:385–407. doi: 10.1146/annurev.genet.40.110405.090423. [DOI] [PubMed] [Google Scholar]
- 40.Romling U, Amikam D. Cyclic di-GMP as a second messenger. Curr Opin Microbiol. 2006;9:218–228. doi: 10.1016/j.mib.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 41.Tamayo R, Pratt JT, Camilli A. Roles of cyclic diguanylate in the regulation of bacterial pathogenesis. Annu Rev Microbiol. 2007;61:131–148. doi: 10.1146/annurev.micro.61.080706.093426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cotter PA, Stibitz S. c-di-GMP-mediated regulation of virulence and biofilm formation. Curr Opin Microbiol. 2007;10:17–23. doi: 10.1016/j.mib.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 43.Mikkelsen H, Ball G, Giraud C, Filloux A. Expression of Pseudomonas aeruginosa CupD fimbrial genes is antagonistically controlled by RcsB and the EAL-containing PvrR response regulators. PLoS One. 2009;4:e6018. doi: 10.1371/journal.pone.0006018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Claret L, Miquel S, Vieille N, Ryjenkov DA, Gomelsky M, et al. The flagellar sigma factor FliA regulates adhesion and invasion of Crohn disease-associated Escherichia coli via a cyclic dimeric GMP-dependent pathway. J Biol Chem. 2007;282:33275–33283. doi: 10.1074/jbc.M702800200. [DOI] [PubMed] [Google Scholar]
- 45.Malone JG, Jaeger T, Spangler C, Ritz D, Spang A, et al. YfiBNR Mediates Cyclic di-GMP Dependent Small Colony Variant Formation and Persistence in Pseudomonas aeruginosa. PLoS Pathog. 2010;6:e1000804. doi: 10.1371/journal.ppat.1000804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ueda A, Wood TK. Connecting quorum sensing, c-di-GMP, Pel polysaccharide, and biofilm formation in Pseudomonas aeruginosa through tyrosine phosphatase TpbA (PA3885). PLoS Pathog. 2009;5:e1000483. doi: 10.1371/journal.ppat.1000483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Amikam D, Galperin MY. PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics. 2006;22:3–6. doi: 10.1093/bioinformatics/bti739. [DOI] [PubMed] [Google Scholar]
- 48.Murakami KS, Darst SA. Bacterial RNA polymerases: the wholo story. Curr Opin Struct Biol. 2003;13:31–39. doi: 10.1016/s0959-440x(02)00005-2. [DOI] [PubMed] [Google Scholar]
- 49.Ebright RH. Transcription activation at Class I CAP-dependent promoters. Mol Microbiol. 1993;8:797–802. doi: 10.1111/j.1365-2958.1993.tb01626.x. [DOI] [PubMed] [Google Scholar]
- 50.Benach J, Swaminathan SS, Tamayo R, Handelman SK, Folta-Stogniew E, et al. The structural basis of cyclic diguanylate signal transduction by PilZ domains. EMBO J. 2007;26:5153–5166. doi: 10.1038/sj.emboj.7601918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ko J, Ryu KS, Kim H, Shin JS, Lee JO, et al. Structure of PP4397 reveals the molecular basis for different c-di-GMP binding modes by Pilz domain proteins. J Mol Biol. 2010;398:97–110. doi: 10.1016/j.jmb.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 52.Ramelot TA, Yee A, Cort JR, Semesi A, Arrowsmith CH, et al. NMR structure and binding studies confirm that PA4608 from Pseudomonas aeruginosa is a PilZ domain and a c-di-GMP binding protein. Proteins. 2007;66:266–271. doi: 10.1002/prot.21199. [DOI] [PubMed] [Google Scholar]
- 53.Schirmer T, Jenal U. Structural and mechanistic determinants of c-di-GMP signalling. Nat Rev Microbiol. 2009;7:724–735. doi: 10.1038/nrmicro2203. [DOI] [PubMed] [Google Scholar]
- 54.Bryson K, McGuffin LJ, Marsden RL, Ward JJ, Sodhi JS, et al. Protein structure prediction servers at University College London. Nucleic Acids Res. 2005;33:W36–38. doi: 10.1093/nar/gki410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sidote DJ, Barbieri CM, Wu T, Stock AM. Structure of the Staphylococcus aureus AgrA LytTR domain bound to DNA reveals a beta fold with an unusual mode of binding. Structure. 2008;16:727–735. doi: 10.1016/j.str.2008.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nikolskaya AN, Galperin MY. A novel type of conserved DNA-binding domain in the transcriptional regulators of the AlgR/AgrA/LytR family. Nucleic Acids Res. 2002;30:2453. doi: 10.1093/nar/30.11.2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Galperin MY. Structural classification of bacterial response regulators: diversity of output domains and domain combinations. J Bacteriol. 2006;188:4169–4182. doi: 10.1128/JB.01887-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Abdelnour A, Arvidson S, Bremell T, Ryden C, Tarkowski A. The accessory gene regulator (agr) controls Staphylococcus aureus virulence in a murine arthritis model. Infect Immun. 1993;61:3879–3885. doi: 10.1128/iai.61.9.3879-3885.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Novick RP. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol Microbiol. 2003;48:1429–1449. doi: 10.1046/j.1365-2958.2003.03526.x. [DOI] [PubMed] [Google Scholar]
- 60.Lizewski SE, Lundberg DS, Schurr MJ. The transcriptional regulator AlgR is essential for Pseudomonas aeruginosa pathogenesis. Infect Immun. 2002;70:6083–6093. doi: 10.1128/IAI.70.11.6083-6093.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McCrary BS, Edmondson SP, Shriver JW. Hyperthermophile protein folding thermodynamics: differential scanning calorimetry and chemical denaturation of Sac7d. J Mol Biol. 1996;264:784–805. doi: 10.1006/jmbi.1996.0677. [DOI] [PubMed] [Google Scholar]
- 62.Robinson H, Gao YG, McCrary BS, Edmondson SP, Shriver JW, et al. The hyperthermophile chromosomal protein Sac7d sharply kinks DNA. Nature. 1998;392:202–205. doi: 10.1038/32455. [DOI] [PubMed] [Google Scholar]
- 63.Krasteva PV, Fong JC, Shikuma NJ, Beyhan S, Navarro MV, et al. Vibrio cholerae VpsT regulates matrix production and motility by directly sensing cyclic di-GMP. Science. 2010;327:866–868. doi: 10.1126/science.1181185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Leduc JL, Roberts GP. Cyclic di-GMP allosterically inhibits the CRP-like protein (Clp) of Xanthomonas axonopodis pv. citri. J Bacteriol. 2009;191:7121–7122. doi: 10.1128/JB.00845-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tao F, He YW, Wu DH, Swarup S, Zhang LH. The cyclic nucleotide monophosphate domain of Xanthomonas campestris global regulator Clp defines a new class of cyclic di-GMP effectors. J Bacteriol. 2010;192:1020–1029. doi: 10.1128/JB.01253-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hickman JW, Harwood CS. Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol Microbiol. 2008;69:376–389. doi: 10.1111/j.1365-2958.2008.06281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee VT, Matewish JM, Kessler JL, Hyodo M, Hayakawa Y, et al. A cyclic-di-GMP receptor required for bacterial exopolysaccharide production. Mol Microbiol. 2007;65:1474–1484. doi: 10.1111/j.1365-2958.2007.05879.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Doran TI. The role of Citrobacter in clinical disease of children: review. Clin Infect Dis. 1999;28:384–394. doi: 10.1086/515106. [DOI] [PubMed] [Google Scholar]
- 69.Ong CL, Ulett GC, Mabbett AN, Beatson SA, Webb RI, et al. Identification of type 3 fimbriae in uropathogenic Escherichia coli reveals a role in biofilm formation. J Bacteriol. 2008;190:1054–1063. doi: 10.1128/JB.01523-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Volz K. Structural conservation in the CheY superfamily. Biochemistry. 1993;32:11741–11753. doi: 10.1021/bi00095a001. [DOI] [PubMed] [Google Scholar]
- 71.Gerstel U, Römling U. The csgD promoter, a control unit for biofilm formation in Salmonella typhimurium. Res Microbiol. 2003;154:659–667. doi: 10.1016/j.resmic.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 72.Hammar M, Arnqvist A, Bian Z, Olsén A, Normark S. Expression of two csg operons is required for production of fibronectin-and congo red-binding curli polymers in Escherichia coli K-12. Mol Microbiol. 1995;18:661–670. doi: 10.1111/j.1365-2958.1995.mmi_18040661.x.. [DOI] [PubMed] [Google Scholar]
- 73.Romling U, Bian Z, Hammar M, Sierralta WD, Normark S. Curli fibers are highly conserved between Salmonella typhimurium and Escherichia coli with respect to operon structure and regulation. J Bacteriol. 1998;180:722–731. doi: 10.1128/jb.180.3.722-731.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Römling U. Molecular biology of cellulose production in bacteria. Res Microbiol. 2002;153:205–212. doi: 10.1016/s0923-2508(02)01316-5. [DOI] [PubMed] [Google Scholar]
- 75.Römling U, Rohde M, Olsén A, Normark S, Reinköster J. AgfD, the checkpoint of multicellular and aggregative behaviour in Salmonella typhimurium regulates at least two independent pathways. Mol Microbiol. 2000;36:10–23. doi: 10.1046/j.1365-2958.2000.01822.x. [DOI] [PubMed] [Google Scholar]
- 76.Donlan RM, Costerton JW. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev. 2002;15:167–193. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol. 2004;2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 78.Grant SG, Jessee J, Bloom FR, Hanahan D. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc Natl Acad Sci U S A. 1990;87:4645–4649. doi: 10.1073/pnas.87.12.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Herring CD, Glasner JD, Blattner FR. Gene replacement without selection: regulated suppression of amber mutations in Escherichia coli. Gene. 2003;311:153–163. doi: 10.1016/s0378-1119(03)00585-7. [DOI] [PubMed] [Google Scholar]
- 80.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cherepanov PP, Wackernagel W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene. 1995;158:9–14. doi: 10.1016/0378-1119(95)00193-a. [DOI] [PubMed] [Google Scholar]
- 82.Chalker AF, Minehart HW, Hughes NJ, Koretke KK, Lonetto MA, et al. Systematic identification of selective essential genes in Helicobacter pylori by genome prioritization and allelic replacement mutagenesis. J Bacteriol. 2001;183:1259–1268. doi: 10.1128/JB.183.4.1259-1268.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Heydorn A, Nielsen A, Hentzer M, Sternberg C, Givskov M, et al. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology. 2000;146:2395–2407. doi: 10.1099/00221287-146-10-2395. [DOI] [PubMed] [Google Scholar]
- 84.Miller JH. Experiments in molecular genetics: Cold Spring Harbour Laboratory, Cold Spring Harbour, N.Y. 1974. pp. 352–355.
- 85.Shaw WV. Chloramphenicol acetyltransferase from chloramphenicol-resistant bacteria. Methods Enzymol. 1975;43:737–755. doi: 10.1016/0076-6879(75)43141-x. [DOI] [PubMed] [Google Scholar]
- 86.Rao F, Pasunooti S, Ng Y, Zhuo W, Lim L, et al. Enzymatic synthesis of c-di-GMP using a thermophilic diguanylate cyclase. Anal Biochem. 2009;389:138–142. doi: 10.1016/j.ab.2009.03.031. [DOI] [PubMed] [Google Scholar]
- 87.Ryjenkov DA, Simm R, Romling U, Gomelsky M. The PilZ domain is a receptor for the second messenger c-di-GMP: the PilZ domain protein YcgR controls motility in enterobacteria. J Biol Chem. 2006;281:30310–30314. doi: 10.1074/jbc.C600179200. [DOI] [PubMed] [Google Scholar]
- 88.Merighi M, Lee VT, Hyodo M, Hayakawa Y, Lory S. The second messenger bis-(3′-5′)-cyclic-GMP and its PilZ domain-containing receptor Alg44 are required for alginate biosynthesis in Pseudomonas aeruginosa. Mol Microbiol. 2007;65:876–895. doi: 10.1111/j.1365-2958.2007.05817.x. [DOI] [PubMed] [Google Scholar]
- 89.Tagami H, Aiba H. A common role of CRP in transcription activation: CRP acts transiently to stimulate events leading to open complex formation at a diverse set of promoters. EMBO J. 1998;17:1759–1767. doi: 10.1093/emboj/17.6.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kovacikova G, Skorupski K. Overlapping binding sites for the virulence gene regulators AphA, AphB and cAMP-CRP at the Vibrio cholerae tcpPH promoter. Mol Microbiol. 2001;41:393–407. doi: 10.1046/j.1365-2958.2001.02518.x. [DOI] [PubMed] [Google Scholar]
- 91.Qi Y, Rao F, Luo Z, Liang ZX. A flavin cofactor-binding PAS domain regulates c-di-GMP synthesis in AxDGC2 from Acetobacter xylinum. Biochemistry. 2009;48:10275–10285. doi: 10.1021/bi901121w. [DOI] [PubMed] [Google Scholar]
- 92.Larkin M, Blackshields G, Brown N, Chenna R, McGettigan P, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 93.Gouet P, Courcelle E, Stuart DI, Metoz F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
- 94.Rao F, Yang Y, Qi Y, Liang ZX. Catalytic mechanism of cyclic di-GMP-specific phosphodiesterase: a study of the EAL domain-containing RocR from Pseudomonas aeruginosa. J Bacteriol. 2008;190:3622–3631. doi: 10.1128/JB.00165-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nat Biotechnol. 1983;1:784–791. [Google Scholar]
- 96.Casadaban MJ. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J Mol Biol. 1976;104:541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- 97.Studier FW, Moffatt BA. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 98.Chang AC, Cohen SN. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bolivar F, Rodriguez RL, Greene PJ, Betlach MC, Heyneker HL, et al. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2:95–113. [PubMed] [Google Scholar]
- 100.de Lorenzo V, Herrero M, Jakubzik U, Timmis KN. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yang J, Tauschek M, Strugnell R, Robins-Browne RM. The H-NS protein represses transcription of the eltAB operon, which encodes heat-labile enterotoxin in enterotoxigenic Escherichia coli, by binding to regions downstream of the promoter. Microbiology. 2005;151:1199–1208. doi: 10.1099/mic.0.27734-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Biofilm formation by K. pneumoniae AJ218. Biofilm formation by K. pneumoniae AJ218 wild-type and isogenic mutants strains +/- empty pACYC184 plasmids. Biofilm formation was determined using the static microtiter plate assay following incubation in M63B1-GCAA minimal media (supplemented with 1% glycerol and 0.3% casamino acids) for 24 h under static conditions. Results are expressed as a percentage of the biofilm produced by the wild-type AJ218 strain, which is set to 100%. All values represent the mean of four replicate sample wells for each strain performed in two independent experiments. The error bars represent the standard deviation.
(TIF)
Type 3 fimbriae expression by K. pneumoniae AJ218. Mannose resistant Klebsiella-like hemagglutination (MR/K HA) by K. pneumoniae AJ218 wild-type and isogenic mutant strains +/- empty pACYC184 plasmids using human erythrocytes. MR/K HA titer is expressed as the lowest concentration (CFU/mL) of bacteria causing a visible agglutination reaction. Values represent the mean of three independent experiments. The error bars represent the standard deviation.
(TIF)
Coomassie-blue stained SDS-PAGE of over-expressed and purified MrkH-8×His (10 µg loaded). The recombinant MrkH-8×His protein (used for EMSA studies) is labeled, which migrates at approximately 28 kDa.
(TIF)
EMSA of the mrkA fragment. The buffers and conditions used in the assay are as described in the Materials and Methods. The 32P-labelled PCR fragment containing the mrkA regulatory region was generated using primer pairs 32P-Px1mrkARev and mrk295F. The mrkA fragment was mixed with varying amounts of either the purified wild-type MrkH-8×His protein (from 0 to 500 nM) in the presence of 200 μM of GTP (left panel) or the purified mutant MrkH(113R-A)-8×His protein (from 0 to 500 nM) in the presence of 200 μM of c-di-GMP (right panel). Following incubation at 30°C for 20 min, the samples were analyzed on native polyacrylamide gels. The unbound DNA bands (F) are marked.
(TIF)
Immunoblot of MrkH-8×His expression. Samples were prepared by sonication followed by centrifugation and supernatant samples were separated by SDS-PAGE. Following transfer, the membrane was probed with anti-His antibody. Shown are E. coli MC4100 strains harboring pGMrkH-8His (wild-type) and pGMrkH(113R-A)-8His (mutant) preparations. E. coli MC4100 was used as the negative control. MrkH-8×His is labeled, which migrates at approximately 28 kDa.
(TIF)
Coomassie-blue stained SDS-PAGE of over-expressed and purified MrkJ-8×His (10 µg loaded). MrkJ-8×His protein (used for HPLC studies) is labeled, which migrates at approximately 29 kDa.
(TIF)
Oligonucleotide primers used in this study.
(DOC)