Abstract
Background
Nitric oxide (NO) exerts powerful physiological effects through guanylate cyclase (GC), a non-mitochondrial enzyme, and through the generation of protein cysteinyl-NO (SNO) adducts— a post-translational modification relevant to mitochondrial biology. A small number of SNO proteins, generated by various mechanisms, are characteristically found in mammalian mitochondria and influence the regulation of oxidative phosphorylation and other aspects of mitochondrial function.
Scope of Review
The principles by which mitochondrial SNO proteins are formed and their actions, independently or collectively with NO binding to heme, iron-sulfur centers, or to glutathione (GSH) are reviewed on a molecular background of SNO-based signal transduction.
Major Conclusions
Mitochondrial SNO-proteins have been demonstrated to inhibit Complex I of the electron transport chain, to modulate mitochondrial reactive oxygen species (ROS) production, influence calcium-dependent opening of the mitochondrial permeability transition pore (MPTP), promote selective importation of mitochondrial protein, and stimulate mitochondrial fission. The ease of reversibility and the affirmation of regulated S-nitros(yl)ating and denitros(yl)ating enzymatic reactions supports hypotheses that SNO regulates the mitochondrion through redox mechanisms. SNO modification of mitochondrial proteins, whether homeostatic or adaptive (physiological), or pathogenic, is an area of active investigation.
General Significance
Mitochondrial SNO proteins are associated with mainly protective, bur soem pathological effects; the former mainly in inflammatory and ischemia/reperfusion syndromes and the latter in neurodegenerative diseases. Experimentally, mitochondrial SNO delivery is also emerging as a potential new area of therapeutics.
Keywords: Metabolism, mitochondria, nitric oxide, respiration, S-nitrosylation, S-nitrosation
Introduction
The mitochondrial nitric oxide (NO) field is still in an early stage even though some 2,000 papers have been published on the subject since the year 2000. The area has generated strong interest mainly for two reasons: first, many fundamental roles for mitochondria have been discovered that extend well beyond the capacity for oxidative phosphorylation [1], and second, a rich NO biochemistry that encompasses modifications of peptides and proteins has been found to be important in mitochondrial structure and function [2]. The latter includes interactions of NO with heme (metal nitrosyl; M-NO) and iron-sulfur centers (Fe-S), nitration of proteins (R-NO2), and the formation of low molecular weight and protein S-nitrosothiols (RSNO), each posing its own set of investigative challenges [3].
These distinct chemistries, stemming from the prototypical activation of guanylate cyclase (GC) [4], a non-mitochondrial enzyme, have important effects on mammalian mitochondrial physiology, pathology, and cell survival [5]. In this paper, the emphasis is on cysteinyl-NO (SNO) proteins [6, 7], which may exist alone or together with NO binding to heme or Fe-S. An appreciation of SNO chemical biology, singly and collectively in the context of the cysteine proteome [8] benefits contemporary investigations of mitochondrial function.
The information on how NO reacts with and regulates various mitochondrial proteins is incomplete, and as discussed later, has generated some controversy about the roles these fascinating organelles play in cellular energy and redox regulation, heme biosynthesis, calcium and iron homeostasis, and apoptosis [9–11]. Moreover, NO comes into play in the process of mitochondrial quality control, which involves more than 1,000 proteins and encompasses biogenesis, proliferation, and autophagy (mitophagy) through integration of nuclear and mitochondrial gene regulatory and assembly mechanisms [11]. These processes regulate the cell’s functional capacity (e.g. mitochondrial volume and phenotype) and impinge on key aspects of cellular stress resistance and senescence for which mitochondria are increasingly recognized to have a role [12].
The ability to trap and measure SNO-adducts in cells and organelles is challenging because SNO proteins are labile, and reversibility of NO ligation is fundamental to SNO signal transduction [13–17]. This has been a problem since the former controversy over SNO-hemoglobin [18]; nonetheless SNO is associated with the functions of more than 100 proteins [19]. The analytical methodology is covered elsewhere in this issue, and here attention is given mainly to its limitations, particularly in the context of active denitrosylating systems involving S-nitrosoglutathione reductase (GSNOR/ADH Class3) and thioredoxin-2 (Trx2). In order to emphasize the signal transduction aspects of this post-translational modification, I have also used the —S-nitrosylation nomenclature (in lieu of S-nitrosation) to describe the covalent addition of NO to RSH (more correctly to thiolate anion, RS−).
Finally, it is well appreciated that SNO signaling overlaps with the non-specific overproduction of both SNO and other reactive nitrogen species (RNS) during nitrosative stress [20]. This too has been a significant issue experimentally because it requires the nuanced definition of conditions and meticulous and often laborious in situ evaluation of mitochondrial function, and therefore remains much in need of further research. In order to be concise, I have chosen here to focus on sources of mitochondrial NO, the mitochondrial SNO targets, their relationship to mitochondrial glutathione (GSH), and mitochondrial SNO protein functionality in health and disease.
Sources of NO in Mitochondria
Three difficult, but often asked questions on NO in mitochondria have been the center of attention: 1) where does the NO come from; 2) how much NO, if any, do mitochondrial proteins contain physiologically; and 3) when does mitochondrial NO become toxic? The answers generally depend on four factors: the type, location, and function(s) of the protein, and the specific modifications to which it is subject. NO modifications are part of the larger cysteine proteome [8, 21, 22] in which cysteine residues, in addition to disulfide and SNO formation, may undergo other modifications. The addition of GSH to cysteine (glutathionylation or glutathiolation) — often following SNO formation — is a protective, reversible modification. Other oxidative modifications include the formation of sulfenic acid, which can be further oxidized to sulfinic acid (S+2) and then to biologically irreversible sulfonic acid (S+4). Protein cysteines also interact with fatty acids, most often palmitate — called S-acylation or palmitoylation, the catalytic formation of fatty acid esters that produce various functional effects [22]. Finally, hydrogen sulfide (H2S) generated endogenously from free L-cysteine can attach another sulfur to cysteine, yielding the protein hydropersulfide (−SSH). Sulfhydration often involves the same cysteines as S-nitrosylation and often activates the protein.
Mitochondrial NO Generation
The issue of where mitochondrial NO comes from has perhaps generated the most intense interest as well as the most controversy in the field. There is little argument that mitochondria contain bioactive NO— they do, as determined by the presence and formation of nitrosyl heme and copper [23], nitrated proteins [24], and S-nitrosothiols [25] as well as by SNO clearance or denitrosylating mechanisms [26].
The first and most widely appreciated effect of NO on mitochondria is its ability to inhibit cytochrome c oxidase (Complex IV), the terminal acceptor of the electron transport chain (ETC) that reduces O2 to water [27–29]. It is thus important to know whether NO is actually generated by mitochondria or diffuses into them, and whether it regulates or simply inhibits respiration. Many investigators have reported that mitochondria synthesize NO from L-arginine [30–39], e.g. by a mitochondrial NO synthase (mtNOS); however, others have maintained that this is a broken cell artifact [40–44]. The issue is important for two reasons: first is enzyme regulation and second is the sizeable superoxide (·O2−) leak from the electron transport chain [45]— as the chemical combination of NO and ·O2− produces highly toxic peroxynitrite anion (ONOO−), which decomposes to NO2 and an OH radical species [46, 47].
For more than 15 years, there have been reports that one of the isoforms of NO synthase (NOS) is resident in mitochondria. NOS was first reported in liver and rat brain mitochondria in 1995 by Bates et al. who detected neuronal NOS (nNOS; NOS1) by immunochemistry [48]. Subsequent groups have reported NO production, calcium-dependent NOS activity, and NOS localization to the inner mitochondrial membrane— often in liver mitochondria [33, 36, 39, 43, 49–52]. Calcium-induced NO release was also reported in cardiac mitochondria of wild-type mice that was absent in nNOS−/− mice [33].
Subsequent efforts to identify a specific gene for mtNOS failed, but NO production in a number of cases has been attributed to nNOS (NOS1) found in association with mitochondria under some, but not all conditions [39]. The nNOS transcript is an α splice variant [51], and the protein is constitutive to the inner membrane [51], despite having no known mitochondrial leader sequence or other importation mechanism. In liver and brain mitochondria of rats, it has been observed that mtNOS is not detectable by nNOS antibodies to the PDZ domain at the N-terminus, but is detectable by antibodies to C-terminal domains [52, 53]. On this basis, cleavage of this domain has been proposed to enable mitochondrial importation and to account for a lower molecular weight of the protein.
Of further interest is that functional nNOS is often detected in association with heart mitochondria [54] and conditional nNOS over-expression in the mouse heart is connected with Hsp90-associated accumulation of mtNOS, decreased ROS production, and a smaller infarct size after ischemia/reperfusion (IR) [55]. Both iNOS (NOS2) and eNOS (NOS3) have also been detected in mitochondria under normal and under pathological conditions [56–60], such as acute inflammation [61, 62].
Functionally, mtNOS has been proposed to regulate mitochondrial O2 utilization by the reversible formation of the NO-Cu+B moiety of cytochrome c oxidase [49, 52], which derives from the observations that nanomolar NO reversibly inhibits the oxidase [2, 29]. Mitochondrial NOS would thus slow O2 consumption and oxidative phosphorylation, for example, through a NOS substrate, co-factor, or calcium-regulated mechanism. However, this hypothesis has not been tested under strictly physiological conditions.
The activity of mtNOS would require a higher O2 concentration in mitochondria than generally occurs [63], assuming a Km for O2 comparable to nNOS, which is two orders of magnitude higher than that of cytochrome oxidase [64]. Also, mitochondrial superoxide production increases at both high [65] and low [66] PO2, and the generation of reactive nitrogen species, such as ONOO−, indiscriminately inactivates proteins, Fe-S clusters, nucleic acids, and lipids [67]. ONOO− reacts rapidly with thiols to produce thiyl radicals or sulfenic acids, which usually react with GSH or other −SH groups to form disulfides [68]. The effects of ONOO− are generally rapid, non-specific, inhibitory, and irreversible [69]. This NO toxicity along with technical concerns about mitochondrial purity, low-specificity NOS activity measurements, and non-optimized controls have helped sustain the mtNOS controversy [42–44].
Given that a number of laboratories have observed mtNOS, one must seek to escape this apparent conceptual corner. There are recognized strategies of targeted NOS translocation or tightly restricted NOS regulation, but mitochondria may also dispose of NO harmlessly, for instance, by exporting it as low molecular weight SNO or by converting it to a metabolite such as nitrite (NO2−). Cytochrome oxidase will consume NO in this way when its redox centers are oxidized and electron flow is available [70]. The inactivation of NO in cells depends on the NO: O2 ratio, and it has been suggested that at low O2 concentrations, when cytochrome oxidase is reduced, NO is diverted towards GC to help activate hypoxic vasodilation [71], thereby improving O2 availability at the mitochondria.
External Sources of Mitochondrial NO
Mitochondrial NOS aside, NO can gain entrance to mitochondrial compartments in other ways. The fact that NO lacks an electronic charge and is soluble in hydrophobic environments allows it to diffuse across biological membranes and signal in a paracrine manner at a short distance from its site of generation [72]. Moreover, in cells and tissues, NO generated by calcium-regulated nNOS or eNOS, or by iNOS induction, or by NO donors significantly affects mitochondrial function, but the extent to which this is physiological, pathological, or pharmacological is open to discussion [73].
Bioactive NO can also enter mitochondria as low molecular weight SNO compounds, such as cysteinyl-NO (CSNO) or nitrosoglutathione (GSNO), or as nitrosylated proteins. Peptide cysteine residues participate in Cys-to-Cys NO transfer or transnitrosation reactions that may involve cytoplasmic-to-mitochondrial communication. Although NO2− can be produced from NO by cytochrome oxidase, the reverse reaction, NO production from NO2− by heme-based nitrite reductase activity [74] might generate NO in hypoxic mitochondria without interfering with O2 utilization. This speculation, however, has not been tested experimentally in mitochondria at physiological nitrite concentrations.
Under some conditions, iNOS associates with mitochondria either because it is produced in the cytosol in high abundance or because it is targeted to the outer mitochondrial membrane for a specific purpose [59, 60, 62]. During sepsis in mice, for instance, much of the iNOS in hepatocytes associates with the outer mitochondrial membrane [62]. Endothelial NOS may also attach to the outer mitochondrial membrane during periods of enzyme induction [56].
Targets of Mitochondrial Protein S-nitrosylation
The effort that has gone into the identification of mitochondrial targets of S-nitrosylation has had variable results, and evidence for SNO functionality derives largely from tissues with high ATP utilization rates, such as the heart, brain, and liver. Attempts to identify the SNO-protein population in intact mitochondria indicate that SNO is present in a small subset of proteins and that roughly 1% of the mitochondrial proteome is available for S-nitrosylation [75]. This suggests that mitochondrial SNO proteins formed under cell stress or by injury, or in response to low levels of exogenous S-nitrosylating agents, are candidates for physiological regulation. It is assumed that identifying and characterizing these targets will inform how S-nitrosylation is involved in mitochondrial homeostasis and when SNO aberrancy— under- or over-production— causes pathology. Moreover, mitochondrial-targeting S-nitrosylating agents have been protective in several disease models [75–78].
It is no surprise that the complement of mitochondrial SNO-proteins detected in NO donor experiments varies with biological preparation, donor type and concentration, tissue PO2, and other factors [79]. The analytical procedures used in most studies, e.g. biotin switch or labeling with fluorescent probes, may not detect low abundance proteins and do not allow specific cysteine residues to be identified, which require more sensitive and specific approaches, such as differential alkylation, to precisely locate the modified cysteines [8]. The most sensitive proteomic screening method available to date involves SNO-DIGE (difference in gel electrophoresis), which detect low concentrations of SNO proteins on 2D gels [80]. A resin-assisted method has also been developed to capture SNO proteins and compare them against simultaneous oxidation of cysteines in the same proteins [81] A list of mitochondrial proteins that undergo SNO formation is provided in Table 1, a few of which are discussed briefly below.
Table 1. Mitochondrial Proteins Sensitive to In Situ S-nitrosylation.
A small number of mitochondrial proteins with critical thiol moieties are known to undergo SNO adduct formation in various species including the mouse, rat, and human. The table lists 25 major SNO proteins and their main functions, accession numbers for the human protein from the Protein Knowledgebase (UniProtKB/Swiss-Prot) website, and reference numbers. Note seven of eight citric acid cycle enzymes and five fatty acid oxidation enzymes have been reported to undergo S-nitrosylation.
| Accession Number | Protein | Function | SNO Effect | Reference |
|---|---|---|---|---|
| Q99798 | Aconitase | Citric acid cycle | Inhibitory | 75, 77, 78,101 |
| O75390 | Citrate synthase (CS) | Citric acid cycle | ? | 78 |
| Q02218 | 2-oxoglutarate dehydrogenase | Citric acid cycle | Inhibitory | 75, 77 |
| P48735 | Isocitrate dehydrogenase | Citric acid cycle | ? | 77, 78 |
| P40926 | Malate dehydrogenase | Citric acid cycle | ? | 76, 77, 78 |
|
Q9P2R7 P53597 |
Succinyl CoA ligase Subunits α and β |
Citric acid cycle | ? | 78 |
| P09622 | Dihydrolipoyl dehydrogenase | PDH complex | ? | 76 |
| P03897 | NADH dehydrogenase subunit 3 | ETC Complex I | Inhibitory | 99 |
| P31040 | Succinate dehydrogenase A (SDHA) | ETC Complex II Citric acid cycle |
? | 77, 78 |
| P31930 | Cytochrome b-c1 complex subunit 1 | ETC Complex III | ? | 77, 78 |
| P49748 | Very long chain acyl-CoA dehydrogenase (VLCAD) | Fatty acid oxidation | ? | 77, 78 |
| P16219 | Short chain acyl-CoA dehydrogenase | Fatty acid oxidation | ? | 77 |
| P23786 | Carnitine palmitoyl transferase 2 | Fatty acid oxidation | ? | 77 |
| P30084 | Enoyl CoA hydratase | Fatty acid oxidation | ? | 77, 78 |
| Q16134 | Electron transferring flavoprotein dehydrogenase | Ubiquinone oxidoreductase | ? | 77, 78 |
| P05091 | Aldehyde dehydrogenase-2 (ALDH2) | Alcohol metabolism | Inhibitory | 100 |
| O15382 | Branched chain amino acid aminotransferase (BCAT2) | Amino acid catabolism | Inhibitory | 110 |
| P10809 | Heat shock protein 60 (Hsp60) | Chaperonin | Activating | 59 |
| P38646 | Heat shock protein 70 (mortalin) | Chaperonin | Activating | 59 |
| O00429 | Dynamin-related protein-1 (Drp1) | Mitochondrial Fission | Activating | 142 |
| Q9UL12 | Sarcosine dehydrogenase | Sarcosine degradation | ? | 76 |
| P54868 | Hydroxymethylglutaryl CoA synthase | Mevalonate biosynthesis | ? | 76 |
| P00505 | Aspartate aminotransferase | Amino acid exchange | ? | 78 |
| P45880 | VDAC2 | Voltage dependent anion channel | ? | 78 |
Complex I and Other ETC Components
Mitochondria utilize four ETC complexes (I-IV) and the ATP synthase (Complex V) to carry out oxidative phosphorylation. Studies of the effects of NO on the ETC actually originated nearly 50 years ago with cytochrome c oxidase [82]. Historically, because its chemical behavior resembles that of O2, NO has been used as a probe, like CO, to inhibit O2 utilization by the oxidase [28]. Brief exposures to low concentrations of NO promptly and reversibly inhibit cytochrome oxidase [83]. NO interacts with both the oxidized and reduced oxidase complex, and depending on the redox states of the four centers, are of two main types; one is competitive with O2 (like CO), while the other is non-competitive [84].
At a physiological PO2, ~60 nM NO is required to inhibit cellular respiration by ~50% [85]. Thus, at a normal PO2, cytochrome oxidase is 50-fold less sensitive to NO than is GC. In order for NO generated physiologically in the cytoplasm to inhibit cytochrome oxidase, GC presumably would be almost fully activated. Hence, whether physiological NO actually regulates respiration at cytochrome oxidase when vascular GC is not already activated is an unresolved question [2, 86, 87]. Nevertheless, a variety of NOS-expressing cells do show NO-mediated suppression of respiration by some mechanism [2, 86–89].
NO has also long been known to inhibit Complex I [90]. In cells deficient in mitochondrial superoxide production, NO inhibits both Complexes I and IV [91, 92]. Unlike Complex IV inhibition, which may remain reversible, Complex I inhibition easily becomes irreversible due to oxidation and nitration of susceptible subunits of the complex [93]. Complex I inhibition by NO also increases as reduced glutathione (GSH) concentration decreases in the cell [91, 94], which provided early evidence of roles for S-nitrosylation and for glutathionylation [91, 93–96].
Mammalian Complex I is composed of 45 different subunits [97], which include a single flavin mononucleotide and eight iron-sulfur groups [98] that catalyze a two-electron transfer from NADH to ubiquinone with the movement of 4H+ across the inner mitochondrial membrane [99]. Complex I is inactivated by SNO in several ways, but in most cases, inactivation can be reversed with dithiothreitol, copper/ascorbate, or by exposure to light, all consistent with a role for S-nitrosylation [93]. Moreover, many SNO compounds reversibly inactivate Complex I even if little or no free NO is available, implying the involvement of transnitrosation [91, 94].
A property of Complex I called the active/deactive (A/D) transition causes a delay in the onset of NADH:ubiquinone oxidoreductase activity or its reverse [100]. Deactive or dormant (D) enzyme is reactivated by stimulation of enzyme turnover [100]. The active (A) form is resistant while the D-form is sensitive to inactivation by SNO [93], but this may be relevant only under pathological conditions, when enzyme turnover is limited by hypoxia or by a high NO: O2 ratio [93, 101]. A critical thiol has been identified in the catalytic region of Complex I by fluorescence labeling and proteomic analysis at Cys-39 of the mitochondrial-encoded ND3 subunit in bovine heart mitochondria [102]. Cys-39 is found in a loop connecting the first and second transmembrane helix of ND3 that may be a specific site of physiological SNO regulation.
Metabolic Pathways
Apart from Complex I, other metabolic enzymes can be inhibited by S-nitrosylation, such as mitochondrial aldehyde dehydrogenase (ALDH2) [103] and aconitase [104]. Using a mitochondria-targeted S-nitrosothiol (MitoSNO) and SNO-DIGE, one study reported S-nitrosylation of eight enzymes of carbohydrate and fatty acid metabolism in rat heart mitochondria [80]. Five are involved in fatty acid β-oxidation including the very-long-chain acyl-CoA dehydrogenase, the short-chain acyl- CoA dehydrogenase, and enoyl-CoA hydratase. Also identified were carnitine palmitoyl transferase 2, which initiates β-oxidation, and the electron-transferring flavoprotein dehydrogenase, which transfers electrons derived from the oxidation of fatty acids to ubiquinone. Three citric acid cycle enzymes were also S-nitrosylated: aconitase, α-ketoglutarate dehydrogenase, and the succinate dehydrogenase flavoprotein (subunit A), a part of Complex II. The activity of three of these eight enzymes— aconitase, α-KGDH and ALDH2— was checked and found to be inhibited by SNO.
The investigators also postulated that decreased Complex I activity by S-nitrosylation may limit cardiac mitochondrial ROS production and calcium overload during ischemia-reperfusion (IR) [94, 105, 106]. They noted that temporary inhibition of the citric acid cycle and β-oxidation may prevent NADH and reduced ubiquinone from building up and creating a burst of ROS on reperfusion that would damage mitochondria [107]. It is also thought that S-nitrosylation may directly prevent irreversible protein inactivation by ROS [78, 81] or by promoting glutathionylation [108–112].
The effects of NO on the cytosolic and mitochondrial isoforms of the branched-chain amino-acid aminotransferases (BCATc and BCATm, respectively), which control the first step in the catabolism of essential branched chain amino acids, have also been explored [113]. Several NO donors including GSNO inactivate both human isoforms in a dose-dependent manner. Low GSNO concentrations cause a time-dependent inhibition of BCAT that correlates with loss of thiol groups. Although GSNO inactivates both, S-nitrosylation of the cytosolic, but not the mitochondrial isoform was found, suggesting differential regulation by NO. Reversal of GSNO-BCAT with GSH alone was limited, but full reactivation was possible with GSH/glutaredoxin, adding support to the physiological denitrosylation hypothesis.
The Mitochondrial Permeability Transition Pore (MPTP)
When mitochondria are overloaded with calcium, the inner mitochondrial membrane becomes permeable to solute and to molecules of <1.5 kDa because of the opening of a non-selective mitochondrial permeability transition pore (MPTP) [114]. Opening of the MPTP is facilitated by elevated phosphate concentrations, depletion of adenine nucleotides, and/or oxidation of critical thiols associated with the pore [115]. Pore opening uncouples oxidative phosphorylation and worsens ATP depletion, calcium homeostasis, and oxidative stress, leading to cell death by necrosis, apoptosis, or by intersecting mechanisms [116]. Pathologically, energy failure, ROS production, and calcium overload are major interactive factors in MPTP opening in IR injury of the heart [115], brain [117], and liver [118], while in the heart, pre-conditioning dampens these triggers and reduces the sensitivity of the MPTP to calcium [119].
Although the behavior of the MPTP is fairly well elucidated, its molecular composition is not. There is a role for cyclophilin-D (CyP-D)— a peptidyl-prolyl cis-trans isomerase involved in calcium regulation— for which hyper-activation most likely provokes a conformational change in inner membrane proteins at the pore site [120]. Also involved in pore regulation, but not a direct constituent, is the adenine nucleotide translocase (ANT) [121], which allows ADP and ATP to inhibit MPTP opening [122]. The mitochondrial phosphate carrier (PiC) has also been implicated in pore formation, and Halestrap has suggested that calcium triggers a conformational change in the PiC that is facilitated by CyP-D [123]. Agents that modulate the MPTP interact with critical thiol moieties on either or both the PiC and the ANT.
The effects of NO on the MPTP are variable, and it is unclear whether SNO plays a role in physiological MPTP regulation [124]. In general, low NO concentrations delay pore opening in response to calcium [38, 125], while higher concentrations behave like strong oxidants that facilitate pore opening [35, 126]. In studies to evaluate how pharmacological manipulation of the MPTP affects ischemic damage, NO was protective, but this involved both direct and indirect mechanisms [127–129]. In the heart, over-expression of iNOS inhibits MPTP opening during IR [130]. In other cases, SNO enhances MPTP opening— most notably in models of disease where the specificity of interventions for the MPTP is low due to SNO effects on other aspects of mitochondrial function [131, 132]. Moreover, pharmacological and perhaps physiological SNO levels have important non-mitochondrial effects that may impact MPTP function, for instance, through altered Ca++ regulation by the sarcoplasmic/endoplasmic reticulum Ca-ATPase (SERCA) [133–135].
Chaperonins
Most mitochondrial proteins are synthesized as cytoplasmic pre-proteins and imported across the outer and inner membrane by translocases and chaperonins that guide the entry, processing, and folding of the peptides [136]. These import systems are crucial for mitochondrial quality control and for mitochondrial biogenesis [137]. NO activates mitochondrial biogenesis [138], which improves the physiological responses to increased ATP demand and to increased turnover of mitochondria [139]. However, the extent to which NO contributes to mitochondrial protein importation is only now being investigated.
In systemic inflammatory states, such as sepsis, a diminished capacity for oxidative phosphorylation from ETC damage occurs in which NO production by iNOS may contribute [140]. Moreover, increased demand for ATP is met by adjusting energy management and by increasing the rates of mitochondrial biogenesis and mitophagy. Mitochondrial biogenesis involves NO and is not fully met in sepsis in iNOS−/− mice, which display impaired hepatic recovery of mtDNA copy number and peak respiration rates [62, 141]. The absence of iNOS in sepsis is associated with failure to increase the levels of mitochondrial transcription factor-A (Tfam) and DNA polymerase gamma (Polγ) during mitochondrial biogenesis [62]. The iNOS binds to the outer mitochondrial membrane and S-nitrosylates heat shock proteins (Hsp) 60 and Hsp70 (mortalin), which are involved in the importation and folding of mitochondrial proteins translated in the cytoplasm [142]. The iNOS specifically regulates Tfam binding to Hsp 60 through S-nitrosylation of Cys-237, which leads to increases in mitochondrial levels of the transcription factor.
Mitochondrial Fission Proteins
Mitochondria undergo synchronized structural modifications— known as fission and fusion— that regulate their numbers and size, and the supply of energy to different parts of the cell [143]. Recent discoveries have indicated that excessive fission can be triggered in neurons by NO-related dysfunction of the fission-inducing dynamin-related protein-1 (Drp1) on the outer mitochondrial membrane [144]. Drp1 contains an N-terminal GTPase domain and a C-terminal GTPase effector domain (GED) involved in homodimer formation and GTPase activation that is necessary for fission [145].
In neurons, NO induces mitochondrial fission, and S-nitrosylation of Drp1 activates both the GTPase and fission. Drp1 contains nine cysteines, each of which has been mutated and the constructs transfected into cells [11, 144, 145]. Of the nine, only Cys-644 mutation in the GED domain decreased SNO-Drp1 formation. In neurons, nNOS activation leads to SNO-Drp1 formation, which is absent in Drp1-Cys-644 mutant cells, but present in the other mutants. Drp-1-Cys-644 mutant over-expression prevented NO-mediated mitochondrial fragmentation while the other mutations did not, indicating that Cys-644 is the critical Drp1 S-nitrosylation site. Further evidence supports that mitochondrial fragmentation contributes to synaptic damage and neuronal death through increased nitrosative and oxidative stress and bioenergetic compromise. This fragmentation process, along with other NO-dependent excitotoxic events, has been associated with neurodegenerative conditions, such as Alzheimer’s disease [146].
Role of the Mitochondrial Glutathione System
NO and Glutathionylation
As mentioned earlier, NO promotes glutathionylation— the reversible post-translational addition of GSH to protein thiols [147–149]. Formation of these mixed disulfides is involved in redox regulation of protein structure and function. Mitochondria contain millimolar GSH, which is acquired largely through organic anion exchangers [150]. The reduced GSH pool in mitochondria is typically high because GSH oxidation to the disulfide (GSSG), for instance, by mitochondrial glutathione peroxidase, leads to prompt reduction by mitochondrial glutathione reductase or to the export of GSSG to the cytosol [151].
Many mitochondrial proteins contain critical thiol residues, and mitochondria contain detectable S-nitrosoglutathione (GSNO), which forms by direct interaction of NO with GSH or by transnitrosation reactions with other S-nitrosylated proteins [152]. Studies of proteins containing reactive sulfhydryls indicate that micromolar NO or GSNO will reversibly generate S-nitrosylated and/or S-glutathionylated forms, lending support to redox-regulation or redox-switch hypotheses [91, 111, 148, 149, 153]. Similar reversibility can be obtained in cells by exposing them to GSNO or other NO donors at low concentrations. This has been demonstrated, for example, for the cytoplasmic glycolytic enzyme, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) [148]. Thiol residues on GAPDH were S-nitrosylated by the NO donor nitrosonium tetrafluoroborate (BF4NO), which inhibited the enzyme activity. This SNO decomposed spontaneously, restoring activity. In cells, GSNO promoted glutathionylation of GAPDH and enzyme inhibition was reversed by reduction with dithiothreitol [148]. Accordingly, glutathione disulfide (GSSG) also reversibly inactivates certain enzymes by S-glutathionylation, e.g. creatine kinase [110]. It has also been suggested that cytochrome c may catalyze GSNO formation well enough to inhibit Complex I [154].
The data on ETC damage by NO and its mitigation by GSH [91, 155] has given rise to the view that S-glutathionylation safeguards mitochondrial proteins from oxidation/nitration and regulates cell metabolism. Glutathionylation blocks cysteine oxidation by preventing the formation of disulfides and higher oxides of sulfur that may be irreparable. In the presence of NO, the GSH content also strongly influences the amount of ONOO− and other oxidants that is detected; for example, GSH depletion renders rat hepatocytes more sensitive to NO toxicity [156]. Protein thiols in mitochondrial membranes are more easily S-nitrosylated by low molecular weight SNO than by NO or ONOO−, and they are reduced by GSH— tending to endure mainly when the mitochondrial GSH pool is oxidized or depleted [94].
The importance of GSH protection is illustrated by the sensitivity of citric acid cycle aconitase to inactivation by ONOO−. Low micromolar ONOO− inactivates aconitase by reducing the Fe-S cluster to an inactive form with depletion of labile iron. The ONOO− concentration for half-inhibition is greatly increased by citrate binding to the active site, and the enzyme is inactivated by the nitration of Tyr-151 and -472 and the oxidation of Cys-126 and -385 to sulfonic acid [69]. The latter cysteine binds the Fe-S cluster and the other residues are nearby, suggesting these modifications disrupt the active site. Aconitase is also reversibly inhibited by glutathionylation, suggesting this as a mode of modulating citric acid cycle activity and/or preventing a permanent failure [69, 104].
There are also differences in the NO-glutathione effect on mitochondrial function in different cell types. For instance, astrocytes display lower sensitivity to NO toxicity than neurons in part because of up-regulation of GSH synthesis [157]. Exposure of primary cortical neurons or astrocytes to a steady flux of NO (~0.25 microM/sec) decreases GSH and increases GSNO and GSSG formation. Dose-dependent oxidations and correlations between cell viability and redox state are observed. Neurons also show a higher sensitivity to NO than do astrocytes due to a lower capacity to recover GSH through glutathione reductase [158].
Glutathione and denitrosylation
Mitochondria also perform enzymatic denitrosylation [26] and deglutathionylation [108, 159], indicating tight regulation of these processes. The mitochondrial GSH-dependent oxidoreductase glutaredoxin-2 (Grx-2) helps maintain redox homeostasis through the efficient catalysis of both mono- and di-thiol reactions [160]. Grx-2 has a high affinity for protein-glutathione disulfides, but also accepts electrons from thioredoxin reductase, allowing it to support mono- and di-thiol detoxification. Compared with cytosolic Grx-1, mitochondrial Grx-2 retains activity after oxidant exposure, does not form inactive disulfide homodimers, and is relatively resistant to S-nitrosylation [161].
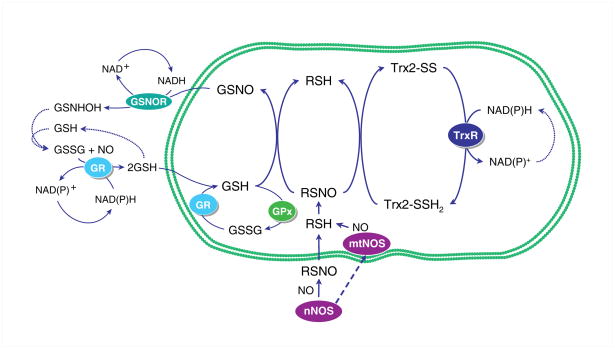
The involvement of GSH in protein denitrosylation enables the attainment of a complete cycle of S-nitrosylation and denitrosylation as shown in Figure 1. Several enzymes can reduce SNO and are thus physiological denitrosylases (see [162]). The prototype is the cytosolic GSNO reductase (GSNOR/ADH Class 3), which uses NADH to convert GSNO to GSNHOH and then to GSSG [163]. The cycle is completed by reduction of GSSG to GSH by glutathione reductase [164]. GSNOR acts on GSNO, but not on SNO proteins, yet affects the equilibrium between GSNO and the SNO-protein pools and helps set SNO protein levels in the cell.
Figure 1.
S-nitrosylation and denitrosylation of mitochondrial proteins with available −SH groups. The transnitrosylation of glutathione (GSH) by S-nitrosylated proteins (RSNO) generates GSNO and the unmodified protein (RSH). The thioredoxin-2 (Trx2) system is shown inside the mitochondrion coupled to protein denitrosylation. Oxidized Trx2 is then reduced by thioredoxin reductase (TrxR2), which consumes NADPH to regenerate active Trx2. The GSNOR cycle is shown in the cytoplasm putatively linked to mitochondrial protein denitrosylation in the context of glutathione recycling (Glutathione peroxidase; GPx and glutathione reductase; GR). GSNO is reduced by GSNOR to generate GSNHOH, which further reacts with GSH to generate GSSG. The cycle is completed by GSSG reduction to GSH by GR.
By comparison, the thioredoxins (Trx1 and Trx2) lack GSNOR’s stringent substrate specificity and thus mediate denitrosylation of many SNO proteins [26]. Following denitrosylation, reduced Trx is regenerated by Trx reductase (TrxR) at the cost of NADPH. Reduced Trx also provides electrons to methionine sulfoxide reductase for repair of methionine sulfoxide residues and to peroxiredoxins for detoxification of H2O2 and ONOO− [164]. A key concept is that the level of protein S-nitrosylation is not determined simply by the rate of SNO generation, but by a local balance among S-nitrosylation, denitrosylation, and -SH protection. Accordingly, protein denitrosylation is an active part of SNO-based signal transduction.
Functionality of Mitochondrial S-nitrosothiols: Physiological Regulation vs. Toxicity
The previous sections summarized the main evidence that mitochondrial S-nitrosothiols play important physiological and adaptive roles in cell stress and that SNO deregulation is involved in the pathogenesis of certain diseases. It should be reiterated that a conceptual difficulty has been a lack of a clear demarcation between primary physiological mechanisms of regulation and secondary responses to NO toxicity for some mitochondrial functions. Several examples have been cited, such as inhibition of Complex I in cardiac IR, S-nitrosylation of Hsp60 in sepsis, and deregulation of mitochondrial fission in neurodegenerative disease. In each case, problems may arise due to multiple inputs to the SNO —on and off mechanisms and the possibility that SNO regulation fails— on or off— in disease.
In the heart, where S-nitrosylation is closely connected to protection from IR injury, it is known that certain pharmacological agents, such as the nitro-vasodilator nitroglycerin, are activated in mitochondria and lead to SNO protein accumulation [162]. This implies a protective role for S-nitrosylation, as seen during preconditioning [165]. The delivery of S-nitrosylating agents to mitochondria is also protective in cardiac IR possibly through the inhibition of ROS generation at Complex I [75]. Other examples include S-nitrosylation of cardiac proteins involved in protective estrogen receptor activation [166] and statin-mediated S-nitrosylation and activation of thioredoxin, resulting in less cellular damage by ROS [167].
The heart can be protected from ischemia by a brief period of ischemic preconditioning (IPC), which increases NO generation [168]. The data suggest that IPC results in protein S-nitrosylation, which contributes to cardiac protection. An increase in mitochondrial SNO protein content in pre-conditioned rat hearts has been postulated to improve cell survival by regulating respiration, redox state, and/or the MPTP [105, 169]. The induction of iNOS may also contribute to the protective effects of IPC through mitochondrial SNO generation [130, 170].
Multiple mitochondrial enzymes that are inhibited by S-nitrosylation, such as Complex I and citric acid cycle enzymes, show increased S-nitrosylation after IPC, while others require an additional S-nitrosylating agent. Exactly how such combinations of SNO proteins protect against cardiomyocyte cell death remains under investigation, but a temporary interruption of respiration may prevent excessive generation of superoxide and ONOO− in early reperfusion and alleviate harmful thiol oxidation that might permanently inactivate metabolic enzymes [81] or permit MPTP opening. A SNO-block at Complex I would prevent reducing equivalents as NADH generated by the citric acid cycle from entering the ETC while oxidation of FADH2 from succinate and fatty acids (the preferred substrate of the heart) could continue. The accumulation of matrix NADH could hypothetically support mitochondrial NADPH generation and mitochondrial antioxidant defenses. If however, S-nitrosylation also blocks pyruvate and fatty acid oxidation [80], oxidative phosphorylation would be interrupted and mitochondrial PO2 would rise. Such deep interference with cellular ATP provision, if widespread, must be reversed quickly in order to avoid cell death and permanent loss of contractile function.
Conclusions
Evidence for a physiological role for SNO formation in the regulation of a small but significant number of mitochondrial proteins is steadily accumulating, but there is still some uncertainty about how SNO is generated, reacts with, and regulates these and perhaps other proteins. Persuasive evidence for SNO regulation of mitochondrial functions comes from studies on inhibition of Complex I and steps in intermediary and fatty acid metabolism, the activation of mitochondrial protein importation, and in neurons, the regulation of mitochondrial fission. The presence of both S-nitrosylating and denitrosylating mechanisms and the readiness of SNO reversibility lends rationale to redox-regulation hypotheses for mitochondria, especially in tissues with high energy requirements. Although there is not yet a cohesive mitochondrial SNO story, the evidence implies that an adaptive —SNO-block protects the organelle under stress, perhaps as an anti-oxidant strategy or as a prelude to mitochondrial fission and/or biogenesis. A fuller understanding of mitochondrial SNO in the future will predictably improve our insights into cell physiology and pathology as well as reveal new avenues of mitochondrial therapy.
Acknowledgments
The author thanks Barry W. Allen, Ph.D. for critically reading the manuscript. Supported by NIH R01 AI064789.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Duchen MR. Mitochondria in health and disease: perspectives on a new mitochondrial biology. Mol Aspects Med. 2004;25:365–451. doi: 10.1016/j.mam.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 2.Brown GC. Nitric oxide and mitochondria. Front Biosci. 2007;12:1024–33. doi: 10.2741/2122. [DOI] [PubMed] [Google Scholar]
- 3.Hetrick EM, Schoenfisch MH. Analytical chemistry of nitric oxide. Annu Rev Anal Chem (Palo Alto Calif) 2009;2:409–33. doi: 10.1146/annurev-anchem-060908-155146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arnold WP, Mittal CK, Katsuki S, Murad F. Nitric oxide activates guanylate cyclase and increases guanosine 3′:5′-cyclic monophosphate levels in various tissue preparations. Proc Natl Acad Sci U S A. 1977;74:3203–7. doi: 10.1073/pnas.74.8.3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Erusalimsky JD, Moncada S. Nitric oxide and mitochondrial signaling: from physiology to pathophysiology. Arterioscler Thromb Vasc Biol. 2007;27:2524–31. doi: 10.1161/ATVBAHA.107.151167. [DOI] [PubMed] [Google Scholar]
- 6.Ignarro LJ, Gruetter CA. Requirement of thiols for activation of coronary arterial guanylate cyclase by glyceryl trinitrate and sodium nitrite: possible involvement of S-nitrosothiols. Biochim Biophys Acta. 1980;631:221–31. doi: 10.1016/0304-4165(80)90297-4. [DOI] [PubMed] [Google Scholar]
- 7.Stamler JS, Simon DI, Osborne JA, Mullins ME, Jaraki O, Michel T, Singel DJ, Loscalzo J. S-nitrosylation of proteins with nitric oxide: synthesis and characterization of biologically active compounds. Proc Natl Acad Sci U S A. 1992;89:444–8. doi: 10.1073/pnas.89.1.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leonard SE, Carroll KS. Chemical ‘omics’ approaches for understanding protein cysteine oxidation in biology. Curr Opin Chem Biol. 2011;15:88–102. doi: 10.1016/j.cbpa.2010.11.012. [DOI] [PubMed] [Google Scholar]
- 9.Baines CP. The cardiac mitochondrion: nexus of stress. Annu Rev Physiol. 2010;72:61–80. doi: 10.1146/annurev-physiol-021909-135929. [DOI] [PubMed] [Google Scholar]
- 10.Wallace DC, Fan W, Procaccio V. Mitochondrial energetics and therapeutics. Annu Rev Pathol. 2010;5:297–348. doi: 10.1146/annurev.pathol.4.110807.092314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakamura T, Lipton SA. Cell death: protein misfolding and neurodegenerative diseases. Apoptosis. 2009;14:455–68. doi: 10.1007/s10495-008-0301-y. [DOI] [PubMed] [Google Scholar]
- 12.Robb EL, Page MM, Stuart JA. Mitochondria, cellular stress resistance, somatic cell depletion and lifespan. Curr Aging Sci. 2009;2:12–27. doi: 10.2174/1874609810902010012. [DOI] [PubMed] [Google Scholar]
- 13.Arnelle DR, Stamler JS. NO+, NO, and NO- donation by S-nitrosothiols: implications for regulation of physiological functions by S-nitrosylation and acceleration of disulfide formation. Arch Biochem Biophys. 1995;318:279–85. doi: 10.1006/abbi.1995.1231. [DOI] [PubMed] [Google Scholar]
- 14.Jaffrey SR, Erdjument-Bromage H, Ferris CD, Tempst P, Snyder SH. Protein S-nitrosylation: a physiological signal for neuronal nitric oxide. Nat Cell Biol. 2001;3:193–7. doi: 10.1038/35055104. [DOI] [PubMed] [Google Scholar]
- 15.Lin TK, Hughes G, Muratovska A, Blaikie FH, Brookes PS, Darley-Usmar V, Smith RA, Murphy MP. Specific modification of mitochondrial protein thiols in response to oxidative stress: a proteomics approach. J Biol Chem. 2002;277:17048–56. doi: 10.1074/jbc.M110797200. [DOI] [PubMed] [Google Scholar]
- 16.Tarpey MM, Wink DA, Grisham MB. Methods for detection of reactive metabolites of oxygen and nitrogen: in vitro and in vivo considerations. Am J Physiol Regul Integr Comp Physiol. 2004;286:R431–44. doi: 10.1152/ajpregu.00361.2003. [DOI] [PubMed] [Google Scholar]
- 17.Mannick JB, Schonhoff CM. Measurement of protein S-nitrosylation during cell signaling. Methods Enzymol. 2008;440:231–42. doi: 10.1016/S0076-6879(07)00814-2. [DOI] [PubMed] [Google Scholar]
- 18.Allen BW, Stamler JS, Piantadosi CA. Hemoglobin, nitric oxide and molecular mechanisms of hypoxic vasodilation. Trends Mol Med. 2009;15:452–60. doi: 10.1016/j.molmed.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Foster MW, Hess DT, Stamler JS. Protein S-nitrosylation in health and disease: a current perspective. Trends Mol Med. 2009;15:391–404. doi: 10.1016/j.molmed.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hausladen A, Fridovich I. Superoxide and peroxynitrite inactivate aconitases, but nitric oxide does not. J Biol Chem. 1994;269:29405–8. [PubMed] [Google Scholar]
- 21.Mustafa AK, Gadalla MM, Sen N, Kim S, Mu W, Gazi SK, Barrow RK, Yang G, Wang R, Snyder SH. H2S signals through protein S-sulfhydration. Sci Signal. 2009;2:ra72. doi: 10.1126/scisignal.2000464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sen N, Snyder SH. Protein modifications involved in neurotransmitter and gasotransmitter signaling. Trends Neurosci. 2010;33:493–502. doi: 10.1016/j.tins.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cooper CE, Torres J, Sharpe MA, Wilson MT. Nitric oxide ejects electrons from the binuclear centre of cytochrome c oxidase by reacting with oxidised copper: a general mechanism for the interaction of copper proteins with nitric oxide? FEBS Lett. 1997;414:281–4. doi: 10.1016/s0014-5793(97)01009-0. [DOI] [PubMed] [Google Scholar]
- 24.Castro L, Demicheli V, Tortora V, Radi R. Mitochondrial protein tyrosine nitration. Free Radic Res. 2010;45:37–52. doi: 10.3109/10715762.2010.516254. [DOI] [PubMed] [Google Scholar]
- 25.Nishikawa M, Sato EF, Kashiba M, Kuroki T, Utsumi K, Inoue M. Role of glutathione in nitric oxide-dependent regulation of energy metabolism in rat hepatoma cells. Hepatology. 1998;27:422–6. doi: 10.1002/hep.510270216. [DOI] [PubMed] [Google Scholar]
- 26.Benhar M, Forrester MT, Hess DT, Stamler JS. Regulated protein denitrosylation by cytosolic and mitochondrial thioredoxins. Science. 2008;320:1050–4. doi: 10.1126/science.1158265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moncada S, Erusalimsky JD. Does nitric oxide modulate mitochondrial energy generation and apoptosis? Nat Rev Mol Cell Biol. 2002;3:214–20. doi: 10.1038/nrm762. [DOI] [PubMed] [Google Scholar]
- 28.Wilson DF, Erecinska M. Ligands of cytochrome c oxidase. Methods Enzymol. 1978;53:191–201. doi: 10.1016/s0076-6879(78)53024-3. [DOI] [PubMed] [Google Scholar]
- 29.Borutaite V, Brown GC. Rapid reduction of nitric oxide by mitochondria, and reversible inhibition of mitochondrial respiration by nitric oxide. Biochem J. 1996;315(Pt 1):295–9. doi: 10.1042/bj3150295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lopez-Figueroa MO, Caamano C, Morano MI, Ronn LC, Akil H, Watson SJ. Direct evidence of nitric oxide presence within mitochondria. Biochem Biophys Res Commun. 2000;272:129–33. doi: 10.1006/bbrc.2000.2748. [DOI] [PubMed] [Google Scholar]
- 31.Giulivi C. Functional implications of nitric oxide produced by mitochondria in mitochondrial metabolism. Biochem J. 1998;332(Pt 3):673–9. doi: 10.1042/bj3320673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giulivi C, Poderoso JJ, Boveris A. Production of nitric oxide by mitochondria. J Biol Chem. 1998;273:11038–43. doi: 10.1074/jbc.273.18.11038. [DOI] [PubMed] [Google Scholar]
- 33.Kanai AJ, Pearce LL, Clemens PR, Birder LA, VanBibber MM, Choi SY, de Groat WC, Peterson J. Identification of a neuronal nitric oxide synthase in isolated cardiac mitochondria using electrochemical detection. Proc Natl Acad Sci U S A. 2001;98:14126–31. doi: 10.1073/pnas.241380298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bustamante J, Bersier G, Badin RA, Cymeryng C, Parodi A, Boveris A. Sequential NO production by mitochondria and endoplasmic reticulum during induced apoptosis. Nitric Oxide. 2002;6:333–41. doi: 10.1006/niox.2001.0420. [DOI] [PubMed] [Google Scholar]
- 35.Piantadosi CA, Tatro LG, Whorton AR. Nitric oxide and differential effects of ATP on mitochondrial permeability transition. Nitric Oxide. 2002;6:45–60. doi: 10.1006/niox.2001.0368. [DOI] [PubMed] [Google Scholar]
- 36.Lacza Z, Snipes JA, Zhang J, Horvath EM, Figueroa JP, Szabo C, Busija DW. Mitochondrial nitric oxide synthase is not eNOS, nNOS or iNOS. Free Radic Biol Med. 2003;35:1217–28. doi: 10.1016/s0891-5849(03)00510-0. [DOI] [PubMed] [Google Scholar]
- 37.Schild L, Reinheckel T, Reiser M, Horn TF, Wolf G, Augustin W. Nitric oxide produced in rat liver mitochondria causes oxidative stress and impairment of respiration after transient hypoxia. Faseb J. 2003;17:2194–201. doi: 10.1096/fj.02-1170com. [DOI] [PubMed] [Google Scholar]
- 38.Leite AC, Oliveira HC, Utino FL, Garcia R, Alberici LC, Fernandes MP, Castilho RF, Vercesi AE. Mitochondria generated nitric oxide protects against permeability transition via formation of membrane protein S-nitrosothiols. Biochim Biophys Acta. 2010;1797:1210–6. doi: 10.1016/j.bbabio.2010.01.034. [DOI] [PubMed] [Google Scholar]
- 39.Ghafourifar P, Parihar MS, Nazarewicz R, Zenebe WJ, Parihar A. Detection assays for determination of mitochondrial nitric oxide synthase activity; advantages and limitations. Methods Enzymol. 2008;440:317–34. doi: 10.1016/S0076-6879(07)00821-X. [DOI] [PubMed] [Google Scholar]
- 40.Tay YM, Lim KS, Sheu FS, Jenner A, Whiteman M, Wong KP, Halliwell B. Do mitochondria make nitric oxide? no? Free Radic Res. 2004;38:591–9. doi: 10.1080/10715760410001694008. [DOI] [PubMed] [Google Scholar]
- 41.Lacza Z, Horn TF, Snipes JA, Zhang J, Roychowdhury S, Horvath EM, Figueroa JP, Kollai M, Szabo C, Busija DW. Lack of mitochondrial nitric oxide production in the mouse brain. J Neurochem. 2004;90:942–51. doi: 10.1111/j.1471-4159.2004.02553.x. [DOI] [PubMed] [Google Scholar]
- 42.Venkatakrishnan P, Nakayasu ES, Almeida IC, Miller RT. Absence of nitric-oxide synthase in sequentially purified rat liver mitochondria. J Biol Chem. 2009;284:19843–55. doi: 10.1074/jbc.M109.003301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lacza Z, Pankotai E, Busija DW. Mitochondrial nitric oxide synthase: current concepts and controversies. Front Biosci. 2009;14:4436–43. doi: 10.2741/3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brookes PS. Mitochondrial nitric oxide synthase. Mitochondrion. 2004;3:187–204. doi: 10.1016/j.mito.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 45.Brand MD. The sites and topology of mitochondrial superoxide production. Exp Gerontol. 2010;45:466–72. doi: 10.1016/j.exger.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xie YW, Wolin MS. Role of nitric oxide and its interaction with superoxide in the suppression of cardiac muscle mitochondrial respiration. Involvement in response to hypoxia/reoxygenation. Circulation. 1996;94:2580–6. doi: 10.1161/01.cir.94.10.2580. [DOI] [PubMed] [Google Scholar]
- 47.Sharpe MA, Cooper CE. Interaction of peroxynitrite with mitochondrial cytochrome oxidase. Catalytic production of nitric oxide and irreversible inhibition of enzyme activity. J Biol Chem. 1998;273:30961–72. doi: 10.1074/jbc.273.47.30961. [DOI] [PubMed] [Google Scholar]
- 48.Bates TE, Loesch A, Burnstock G, Clark JB. Immunocytochemical evidence for a mitochondrially located nitric oxide synthase in brain and liver. Biochem Biophys Res Commun. 1995;213:896–900. doi: 10.1006/bbrc.1995.2213. [DOI] [PubMed] [Google Scholar]
- 49.Ghafourifar P, Richter C. Nitric oxide synthase activity in mitochondria. FEBS Lett. 1997;418:291–6. doi: 10.1016/s0014-5793(97)01397-5. [DOI] [PubMed] [Google Scholar]
- 50.Tatoyan A, Giulivi C. Purification and characterization of a nitric-oxide synthase from rat liver mitochondria. J Biol Chem. 1998;273:11044–8. doi: 10.1074/jbc.273.18.11044. [DOI] [PubMed] [Google Scholar]
- 51.Elfering SL, Sarkela TM, Giulivi C. Biochemistry of mitochondrial nitric-oxide synthase. J Biol Chem. 2002;277:38079–86. doi: 10.1074/jbc.M205256200. [DOI] [PubMed] [Google Scholar]
- 52.Finocchietto PV, Franco MC, Holod S, Gonzalez AS, Converso DP, Arciuch VG, Serra MP, Poderoso JJ, Carreras MC. Mitochondrial nitric oxide synthase: a masterpiece of metabolic adaptation, cell growth, transformation, and death. Exp Biol Med (Maywood) 2009;234:1020–8. doi: 10.3181/0902-MR-81. [DOI] [PubMed] [Google Scholar]
- 53.Riobo NA, Melani M, Sanjuan N, Fiszman ML, Gravielle MC, Carreras MC, Cadenas E, Poderoso JJ. The modulation of mitochondrial nitric-oxide synthase activity in rat brain development. J Biol Chem. 2002;277:42447–55. doi: 10.1074/jbc.M204580200. [DOI] [PubMed] [Google Scholar]
- 54.Dedkova EN, Blatter LA. Characteristics and function of cardiac mitochondrial nitric oxide synthase. J Physiol. 2009;587:851–72. doi: 10.1113/jphysiol.2008.165423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Burkard N, Williams T, Czolbe M, Blomer N, Panther F, Link M, Fraccarollo D, Widder JD, Hu K, Han H, Hofmann U, Frantz S, Nordbeck P, Bulla J, Schuh K, Ritter O. Conditional overexpression of neuronal nitric oxide synthase is cardioprotective in ischemia/reperfusion. Circulation. 2010;122:1588–603. doi: 10.1161/CIRCULATIONAHA.109.933630. [DOI] [PubMed] [Google Scholar]
- 56.Gao S, Chen J, Brodsky SV, Huang H, Adler S, Lee JH, Dhadwal N, Cohen-Gould L, Gross SS, Goligorsky MS. Docking of endothelial nitric oxide synthase (eNOS) to the mitochondrial outer membrane: a pentabasic amino acid sequence in the autoinhibitory domain of eNOS targets a proteinase K-cleavable peptide on the cytoplasmic face of mitochondria. J Biol Chem. 2004;279:15968–74. doi: 10.1074/jbc.M308504200. [DOI] [PubMed] [Google Scholar]
- 57.Gonzales GF, Chung FA, Miranda S, Valdez LB, Zaobornyj T, Bustamante J, Boveris A. Heart mitochondrial nitric oxide synthase is upregulated in male rats exposed to high altitude (4,340 m) Am J Physiol Heart Circ Physiol. 2005;288:H2568–73. doi: 10.1152/ajpheart.00812.2004. [DOI] [PubMed] [Google Scholar]
- 58.Chen K, Northington FJ, Martin LJ. Inducible nitric oxide synthase is present in motor neuron mitochondria and Schwann cells and contributes to disease mechanisms in ALS mice. Brain Struct Funct. 2010;214:219–34. doi: 10.1007/s00429-009-0226-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zanella B, Giordano E, Muscari C, Zini M, Guarnieri C. Nitric oxide synthase activity in rat cardiac mitochondria. Basic Res Cardiol. 2004;99:159–64. doi: 10.1007/s00395-003-0454-3. [DOI] [PubMed] [Google Scholar]
- 60.Buchwalow IB, Schulze W, Karczewski P, Kostic MM, Wallukat G, Morwinski R, Krause EG, Muller J, Paul M, Slezak J, Luft FC, Haller H. Inducible nitric oxide synthase in the myocard. Mol Cell Biochem. 2001;217:73–82. doi: 10.1023/a:1007286602865. [DOI] [PubMed] [Google Scholar]
- 61.Lopez LC, Escames G, Tapias V, Utrilla P, Leon J, Acuna-Castroviejo D. Identification of an inducible nitric oxide synthase in diaphragm mitochondria from septic mice: its relation with mitochondrial dysfunction and prevention by melatonin. Int J Biochem Cell Biol. 2006;38:267–78. doi: 10.1016/j.biocel.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 62.Suliman HB, Babiker A, Withers CM, Sweeney TE, Carraway MS, Tatro LG, Bartz RR, Welty-Wolf KE, Piantadosi CA. Nitric oxide synthase-2 regulates mitochondrial Hsp60 chaperone function during bacterial peritonitis in mice. Free Radic Biol Med. 2010;48:736–46. doi: 10.1016/j.freeradbiomed.2009.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wilson DF, Erecinska M, Drown C, Silver IA. Effect of oxygen tension on cellular energetics. Am J Physiol. 1977;233:C135–40. doi: 10.1152/ajpcell.1977.233.5.C135. [DOI] [PubMed] [Google Scholar]
- 64.Rengasamy A, Johns RA. Characterization of endothelium-derived relaxing factor/nitric oxide synthase from bovine cerebellum and mechanism of modulation by high and low oxygen tensions. J Pharmacol Exp Ther. 1991;259:310–6. [PubMed] [Google Scholar]
- 65.Sanders SP, Zweier JL, Kuppusamy P, Harrison SJ, Bassett DJ, Gabrielson EW, Sylvester JT. Hyperoxic sheep pulmonary microvascular endothelial cells generate free radicals via mitochondrial electron transport. J Clin Invest. 1993;91:46–52. doi: 10.1172/JCI116198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Duranteau J, Chandel NS, Kulisz A, Shao Z, Schumacker PT. Intracellular signaling by reactive oxygen species during hypoxia in cardiomyocytes. J Biol Chem. 1998;273:11619–24. doi: 10.1074/jbc.273.19.11619. [DOI] [PubMed] [Google Scholar]
- 67.Poderoso JJ. The formation of peroxynitrite in the applied physiology of mitochondrial nitric oxide. Arch Biochem Biophys. 2009;484:214–20. doi: 10.1016/j.abb.2008.12.020. [DOI] [PubMed] [Google Scholar]
- 68.Radi R, Beckman JS, Bush KM, Freeman BA. Peroxynitrite oxidation of sulfhydryls. The cytotoxic potential of superoxide and nitric oxide. J Biol Chem. 1991;266:4244–50. [PubMed] [Google Scholar]
- 69.Han D, Canali R, Garcia J, Aguilera R, Gallaher TK, Cadenas E. Sites and mechanisms of aconitase inactivation by peroxynitrite: modulation by citrate and glutathione. Biochemistry. 2005;44:11986–96. doi: 10.1021/bi0509393. [DOI] [PubMed] [Google Scholar]
- 70.Unitt DC, Hollis VS, Palacios-Callender M, Frakich N, Moncada S. Inactivation of nitric oxide by cytochrome c oxidase under steady-state oxygen conditions. Biochim Biophys Acta. 2010;1797:371–7. doi: 10.1016/j.bbabio.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 71.Palacios-Callender M, Hollis V, Mitchison M, Frakich N, Unitt D, Moncada S. Cytochrome c oxidase regulates endogenous nitric oxide availability in respiring cells: a possible explanation for hypoxic vasodilation. Proc Natl Acad Sci U S A. 2007;104:18508–13. doi: 10.1073/pnas.0709440104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lancaster JR., Jr A tutorial on the diffusibility and reactivity of free nitric oxide. Nitric Oxide. 1997;1:18–30. doi: 10.1006/niox.1996.0112. [DOI] [PubMed] [Google Scholar]
- 73.Hill BG, Dranka BP, Bailey SM, Lancaster JR, Jr, Darley-Usmar VM. What part of NO don’t you understand? Some answers to the cardinal questions in nitric oxide biology. J Biol Chem. 2010;285:19699–704. doi: 10.1074/jbc.R110.101618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gladwin MT, Kim-Shapiro DB. The functional nitrite reductase activity of the heme-globins. Blood. 2008;112:2636–47. doi: 10.1182/blood-2008-01-115261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Prime TA, Blaikie FH, Evans C, Nadtochiy SM, James AM, Dahm CC, Vitturi DA, Patel RP, Hiley CR, Abakumova I, Requejo R, Chouchani ET, Hurd TR, Garvey JF, Taylor CT, Brookes PS, Smith RA, Murphy MP. A mitochondria-targeted S-nitrosothiol modulates respiration, nitrosates thiols, and protects against ischemia-reperfusion injury. Proc Natl Acad Sci U S A. 2009;106:10764–9. doi: 10.1073/pnas.0903250106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Konorev EA, Joseph J, Tarpey MM, Kalyanaraman B. The mechanism of cardioprotection by S-nitrosoglutathione monoethyl ester in rat isolated heart during cardioplegic ischaemic arrest. Br J Pharmacol. 1996;119:511–8. doi: 10.1111/j.1476-5381.1996.tb15701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chacko BK, Reily C, Srivastava A, Johnson MS, Ye Y, Ulasova E, Agarwal A, Zinn KR, Murphy MP, Kalyanaraman B, Darley-Usmar V. Prevention of diabetic nephropathy in Ins2(+/)(AkitaJ) mice by the mitochondria-targeted therapy MitoQ. Biochem J. 2010;432:9–19. doi: 10.1042/BJ20100308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sun J, Morgan M, Shen RF, Steenbergen C, Murphy E. Preconditioning results in S-nitrosylation of proteins involved in regulation of mitochondrial energetics and calcium transport. Circ Res. 2007;101:1155–63. doi: 10.1161/CIRCRESAHA.107.155879. [DOI] [PubMed] [Google Scholar]
- 79.Foster MW, Stamler JS. New insights into protein S-nitrosylation. Mitochondria as a model system. J Biol Chem. 2004;279:25891–7. doi: 10.1074/jbc.M313853200. [DOI] [PubMed] [Google Scholar]
- 80.Chouchani ET, Hurd TR, Nadtochiy SM, Brookes PS, Fearnley IM, Lilley KS, Smith RA, Murphy MP. Identification of S-nitrosated mitochondrial proteins by S-nitrosothiol difference in gel electrophoresis (SNO-DIGE): implications for the regulation of mitochondrial function by reversible S-nitrosation. Biochem J. 2010;430:49–59. doi: 10.1042/BJ20100633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kohr MJ, Sun J, Aponte A, Wang G, Gucek M, Murphy E, Steenbergen C. Simultaneous Measurement of Protein Oxidation and S-Nitrosylation During Preconditioning and Ischemia/Reperfusion Injury With Resin-Assisted Capture. Circ Res. 2010 doi: 10.1161/CIRCRESAHA.110.232173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gibson QH, Greenwood C. Kinetic Evidence for a Short Lived Intermediate in the Oxidation of Cytochrome C Oxidase by Molecular Oxygen. J Biol Chem. 1965;240:957–8. [PubMed] [Google Scholar]
- 83.Cleeter MW, Cooper JM, Darley-Usmar VM, Moncada S, Schapira AH. Reversible inhibition of cytochrome c oxidase, the terminal enzyme of the mitochondrial respiratory chain, by nitric oxide. Implications for neurodegenerative diseases. FEBS Lett. 1994;345:50–4. doi: 10.1016/0014-5793(94)00424-2. [DOI] [PubMed] [Google Scholar]
- 84.Cooper CE, Brown GC. The inhibition of mitochondrial cytochrome oxidase by the gases carbon monoxide, nitric oxide, hydrogen cyanide and hydrogen sulfide: chemical mechanism and physiological significance. J Bioenerg Biomembr. 2008;40:533–9. doi: 10.1007/s10863-008-9166-6. [DOI] [PubMed] [Google Scholar]
- 85.Brown GC, Cooper CE. Nanomolar concentrations of nitric oxide reversibly inhibit synaptosomal respiration by competing with oxygen at cytochrome oxidase. FEBS Lett. 1994;356:295–8. doi: 10.1016/0014-5793(94)01290-3. [DOI] [PubMed] [Google Scholar]
- 86.Brown GC, Borutaite V. Nitric oxide and mitochondrial respiration in the heart. Cardiovasc Res. 2007;75:283–90. doi: 10.1016/j.cardiores.2007.03.022. [DOI] [PubMed] [Google Scholar]
- 87.Cooper CE, Giulivi C. Nitric oxide regulation of mitochondrial oxygen consumption II: Molecular mechanism and tissue physiology. Am J Physiol Cell Physiol. 2007;292:C1993–2003. doi: 10.1152/ajpcell.00310.2006. [DOI] [PubMed] [Google Scholar]
- 88.Clementi E, Brown GC, Foxwell N, Moncada S. On the mechanism by which vascular endothelial cells regulate their oxygen consumption. Proc Natl Acad Sci U S A. 1999;96:1559–62. doi: 10.1073/pnas.96.4.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Leavesley HB, Li L, Prabhakaran K, Borowitz JL, Isom GE. Interaction of cyanide and nitric oxide with cytochrome c oxidase: implications for acute cyanide toxicity. Toxicol Sci. 2008;101:101–11. doi: 10.1093/toxsci/kfm254. [DOI] [PubMed] [Google Scholar]
- 90.Stuehr DJ, Nathan CF. Nitric oxide. A macrophage product responsible for cytostasis and respiratory inhibition in tumor target cells. J Exp Med. 1989;169:1543–55. doi: 10.1084/jem.169.5.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Clementi E, Brown GC, Feelisch M, Moncada S. Persistent inhibition of cell respiration by nitric oxide: crucial role of S-nitrosylation of mitochondrial complex I and protective action of glutathione. Proc Natl Acad Sci U S A. 1998;95:7631–6. doi: 10.1073/pnas.95.13.7631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Orsi A, Beltran B, Clementi E, Hallen K, Feelisch M, Moncada S. Continuous exposure to high concentrations of nitric oxide leads to persistent inhibition of oxygen consumption by J774 cells as well as extraction of oxygen by the extracellular medium. Biochem J. 2000;346(Pt 2):407–12. [PMC free article] [PubMed] [Google Scholar]
- 93.Galkin A, Moncada S. S-nitrosation of mitochondrial complex I depends on its structural conformation. J Biol Chem. 2007;282:37448–53. doi: 10.1074/jbc.M707543200. [DOI] [PubMed] [Google Scholar]
- 94.Dahm CC, Moore K, Murphy MP. Persistent S-nitrosation of complex I and other mitochondrial membrane proteins by S-nitrosothiols but not nitric oxide or peroxynitrite: implications for the interaction of nitric oxide with mitochondria. J Biol Chem. 2006;281:10056–65. doi: 10.1074/jbc.M512203200. [DOI] [PubMed] [Google Scholar]
- 95.Borutaite V, Brown GC. S-nitrosothiol inhibition of mitochondrial complex I causes a reversible increase in mitochondrial hydrogen peroxide production. Biochim Biophys Acta. 2006;1757:562–6. doi: 10.1016/j.bbabio.2006.02.014. [DOI] [PubMed] [Google Scholar]
- 96.Burwell LS, Nadtochiy SM, Tompkins AJ, Young S, Brookes PS. Direct evidence for S-nitrosation of mitochondrial complex I. Biochem J. 2006;394:627–34. doi: 10.1042/BJ20051435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Carroll J, Fearnley IM, Skehel JM, Shannon RJ, Hirst J, Walker JE. Bovine complex I is a complex of 45 different subunits. J Biol Chem. 2006;281:32724–7. doi: 10.1074/jbc.M607135200. [DOI] [PubMed] [Google Scholar]
- 98.Ohnishi T. Iron-sulfur clusters/semiquinones in complex I. Biochim Biophys Acta. 1998;1364:186–206. doi: 10.1016/s0005-2728(98)00027-9. [DOI] [PubMed] [Google Scholar]
- 99.Di Virgilio F, Azzone GF. Activation of site I redox-driven H+ pump by exogenous quinones in intact mitochondria. J Biol Chem. 1982;257:4106–13. [PubMed] [Google Scholar]
- 100.Vinogradov AD. Catalytic properties of the mitochondrial NADH-ubiquinone oxidoreductase (complex I) and the pseudo-reversible active/inactive enzyme transition. Biochim Biophys Acta. 1998;1364:169–85. doi: 10.1016/s0005-2728(98)00026-7. [DOI] [PubMed] [Google Scholar]
- 101.Taylor CT, Moncada S. Nitric oxide, cytochrome C oxidase, and the cellular response to hypoxia. Arterioscler Thromb Vasc Biol. 2010;30:643–7. doi: 10.1161/ATVBAHA.108.181628. [DOI] [PubMed] [Google Scholar]
- 102.Galkin A, Meyer B, Wittig I, Karas M, Schagger H, Vinogradov A, Brandt U. Identification of the mitochondrial ND3 subunit as a structural component involved in the active/deactive enzyme transition of respiratory complex I. J Biol Chem. 2008;283:20907–13. doi: 10.1074/jbc.M803190200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Moon KH, Kim BJ, Song BJ. Inhibition of mitochondrial aldehyde dehydrogenase by nitric oxide-mediated S-nitrosylation. FEBS Lett. 2005;579:6115–20. doi: 10.1016/j.febslet.2005.09.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tortora V, Quijano C, Freeman B, Radi R, Castro L. Mitochondrial aconitase reaction with nitric oxide, S-nitrosoglutathione, and peroxynitrite: mechanisms and relative contributions to aconitase inactivation. Free Radic Biol Med. 2007;42:1075–88. doi: 10.1016/j.freeradbiomed.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 105.Nadtochiy SM, Burwell LS, Brookes PS. Cardioprotection and mitochondrial S-nitrosation: effects of S-nitroso-2-mercaptopropionyl glycine (SNO-MPG) in cardiac ischemia-reperfusion injury. J Mol Cell Cardiol. 2007;42:812–25. doi: 10.1016/j.yjmcc.2007.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Galkin A, Abramov AY, Frakich N, Duchen MR, Moncada S. Lack of oxygen deactivates mitochondrial complex I: implications for ischemic injury? J Biol Chem. 2009;284:36055–61. doi: 10.1074/jbc.M109.054346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Murphy MP. How mitochondria produce reactive oxygen species. Biochem J. 2009;417:1–13. doi: 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Beer SM, Taylor ER, Brown SE, Dahm CC, Costa NJ, Runswick MJ, Murphy MP. Glutaredoxin 2 catalyzes the reversible oxidation and glutathionylation of mitochondrial membrane thiol proteins: implications for mitochondrial redox regulation and antioxidant DEFENSE. J Biol Chem. 2004;279:47939–51. doi: 10.1074/jbc.M408011200. [DOI] [PubMed] [Google Scholar]
- 109.Requejo R, Hurd TR, Costa NJ, Murphy MP. Cysteine residues exposed on protein surfaces are the dominant intramitochondrial thiol and may protect against oxidative damage. Febs J. 2010;277:1465–80. doi: 10.1111/j.1742-4658.2010.07576.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Reddy S, Jones AD, Cross CE, Wong PS, Van Der Vliet A. Inactivation of creatine kinase by S-glutathionylation of the active-site cysteine residue. Biochem J. 2000;347(Pt 3):821–7. [PMC free article] [PubMed] [Google Scholar]
- 111.Hurd TR, Costa NJ, Dahm CC, Beer SM, Brown SE, Filipovska A, Murphy MP. Glutathionylation of mitochondrial proteins. Antioxid Redox Signal. 2005;7:999–1010. doi: 10.1089/ars.2005.7.999. [DOI] [PubMed] [Google Scholar]
- 112.Mieyal JJ, Gallogly MM, Qanungo S, Sabens EA, Shelton MD. Molecular mechanisms and clinical implications of reversible protein S-glutathionylation. Antioxid Redox Signal. 2008;10:1941–88. doi: 10.1089/ars.2008.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Coles SJ, Easton P, Sharrod H, Hutson SM, Hancock J, Patel VB, Conway ME. S-Nitrosoglutathione inactivation of the mitochondrial and cytosolic BCAT proteins: S-nitrosation and S-thiolation. Biochemistry. 2009;48:645–56. doi: 10.1021/bi801805h. [DOI] [PubMed] [Google Scholar]
- 114.Halestrap AP. What is the mitochondrial permeability transition pore? J Mol Cell Cardiol. 2009;46:821–31. doi: 10.1016/j.yjmcc.2009.02.021. [DOI] [PubMed] [Google Scholar]
- 115.Halestrap AP, Pasdois P. The role of the mitochondrial permeability transition pore in heart disease. Biochim Biophys Acta. 2009;1787:1402–15. doi: 10.1016/j.bbabio.2008.12.017. [DOI] [PubMed] [Google Scholar]
- 116.Lemasters JJ, Theruvath TP, Zhong Z, Nieminen AL. Mitochondrial calcium and the permeability transition in cell death. Biochim Biophys Acta. 2009;1787:1395–401. doi: 10.1016/j.bbabio.2009.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sims NR, Muyderman H. Mitochondria, oxidative metabolism and cell death in stroke. Biochim Biophys Acta. 2010;1802:80–91. doi: 10.1016/j.bbadis.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 118.Pediaditakis P, Kim JS, He L, Zhang X, Graves LM, Lemasters JJ. Inhibition of the mitochondrial permeability transition by protein kinase A in rat liver mitochondria and hepatocytes. Biochem J. 2010;431:411–21. doi: 10.1042/BJ20091741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Murphy E, Steenbergen C. What makes the mitochondria a killer? Can we condition them to be less destructive? Biochim Biophys Acta. 2010 doi: 10.1016/j.bbamcr.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Elrod JW, Wong R, Mishra S, Vagnozzi RJ, Sakthievel B, Goonasekera SA, Karch J, Gabel S, Farber J, Force T, Brown JH, Murphy E, Molkentin JD. Cyclophilin D controls mitochondrial pore-dependent Ca(2+) exchange, metabolic flexibility, and propensity for heart failure in mice. J Clin Invest. 2010;120:3680–7. doi: 10.1172/JCI43171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kokoszka JE, Waymire KG, Levy SE, Sligh JE, Cai J, Jones DP, MacGregor GR, Wallace DC. The ADP/ATP translocator is not essential for the mitochondrial permeability transition pore. Nature. 2004;427:461–5. doi: 10.1038/nature02229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Connern CP, Halestrap AP. Purification and N-terminal sequencing of peptidyl-prolyl cis-trans-isomerase from rat liver mitochondrial matrix reveals the existence of a distinct mitochondrial cyclophilin. Biochem J. 1992;284(Pt 2):381–5. doi: 10.1042/bj2840381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Leung AW, Varanyuwatana P, Halestrap AP. The mitochondrial phosphate carrier interacts with cyclophilin D and may play a key role in the permeability transition. J Biol Chem. 2008;283:26312–23. doi: 10.1074/jbc.M805235200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Vieira H, Kroemer G. Mitochondria as targets of apoptosis regulation by nitric oxide. IUBMB Life. 2003;55:613–6. doi: 10.1080/15216540310001639652. [DOI] [PubMed] [Google Scholar]
- 125.Brookes PS, Salinas EP, Darley-Usmar K, Eiserich JP, Freeman BA, Darley-Usmar VM, Anderson PG. Concentration-dependent effects of nitric oxide on mitochondrial permeability transition and cytochrome c release. J Biol Chem. 2000;275:20474–9. doi: 10.1074/jbc.M001077200. [DOI] [PubMed] [Google Scholar]
- 126.Borutaite V, Morkuniene R, Brown GC. Nitric oxide donors, nitrosothiols and mitochondrial respiration inhibitors induce caspase activation by different mechanisms. FEBS Lett. 2000;467:155–9. doi: 10.1016/s0014-5793(00)01140-6. [DOI] [PubMed] [Google Scholar]
- 127.Kim JS, Ohshima S, Pediaditakis P, Lemasters JJ. Nitric oxide: a signaling molecule against mitochondrial permeability transition- and pH-dependent cell death after reperfusion. Free Radic Biol Med. 2004;37:1943–50. doi: 10.1016/j.freeradbiomed.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 128.Wang G, Liem DA, Vondriska TM, Honda HM, Korge P, Pantaleon DM, Qiao X, Wang Y, Weiss JN, Ping P. Nitric oxide donors protect murine myocardium against infarction via modulation of mitochondrial permeability transition. Am J Physiol Heart Circ Physiol. 2005;288:H1290–5. doi: 10.1152/ajpheart.00796.2004. [DOI] [PubMed] [Google Scholar]
- 129.Kim JS, Ohshima S, Pediaditakis P, Lemasters JJ. Nitric oxide protects rat hepatocytes against reperfusion injury mediated by the mitochondrial permeability transition. Hepatology. 2004;39:1533–43. doi: 10.1002/hep.20197. [DOI] [PubMed] [Google Scholar]
- 130.West MB, Rokosh G, Obal D, Velayutham M, Xuan YT, Hill BG, Keith RJ, Schrader J, Guo Y, Conklin DJ, Prabhu SD, Zweier JL, Bolli R, Bhatnagar A. Cardiac myocyte-specific expression of inducible nitric oxide synthase protects against ischemia/reperfusion injury by preventing mitochondrial permeability transition. Circulation. 2008;118:1970–8. doi: 10.1161/CIRCULATIONAHA.108.791533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Jekabsone A, Dapkunas Z, Brown GC, Borutaite V. S-nitrosothiol-induced rapid cytochrome c release, caspase activation and mitochondrial permeability transition in perfused heart. Biochem Pharmacol. 2003;66:1513–9. doi: 10.1016/s0006-2952(03)00506-9. [DOI] [PubMed] [Google Scholar]
- 132.Kindler DD, Thiffault C, Solenski NJ, Dennis J, Kostecki V, Jenkins R, Keeney PM, Bennett JP., Jr Neurotoxic nitric oxide rapidly depolarizes and permeabilizes mitochondria by dynamically opening the mitochondrial transition pore. Mol Cell Neurosci. 2003;23:559–73. doi: 10.1016/s1044-7431(03)00074-5. [DOI] [PubMed] [Google Scholar]
- 133.Adachi T. Modulation of vascular sarco/endoplasmic reticulum calcium ATPase in cardiovascular pathophysiology. Adv Pharmacol. 2010;59:165–95. doi: 10.1016/S1054-3589(10)59006-9. [DOI] [PubMed] [Google Scholar]
- 134.Adachi T, Weisbrod RM, Pimentel DR, Ying J, Sharov VS, Schoneich C, Cohen RA. S-Glutathiolation by peroxynitrite activates SERCA during arterial relaxation by nitric oxide. Nat Med. 2004;10:1200–7. doi: 10.1038/nm1119. [DOI] [PubMed] [Google Scholar]
- 135.Manevich Y, Townsend DM, Hutchens S, Tew KD. Diazeniumdiolate mediated nitrosative stress alters nitric oxide homeostasis through intracellular calcium and S-glutathionylation of nitric oxide synthetase. PLoS One. 2010;5:e14151. doi: 10.1371/journal.pone.0014151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.van der Laan M, Hutu DP, Rehling P. On the mechanism of preprotein import by the mitochondrial presequence translocase. Biochim Biophys Acta. 2010;1803:732–9. doi: 10.1016/j.bbamcr.2010.01.013. [DOI] [PubMed] [Google Scholar]
- 137.Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N. Importing mitochondrial proteins: machineries and mechanisms. Cell. 2009;138:628–44. doi: 10.1016/j.cell.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Nisoli E, Clementi E, Paolucci C, Cozzi V, Tonello C, Sciorati C, Bracale R, Valerio A, Francolini M, Moncada S, Carruba MO. Mitochondrial biogenesis in mammals: the role of endogenous nitric oxide. Science. 2003;299:896–9. doi: 10.1126/science.1079368. [DOI] [PubMed] [Google Scholar]
- 139.Nisoli E, Carruba MO. Nitric oxide and mitochondrial biogenesis. J Cell Sci. 2006;119:2855–62. doi: 10.1242/jcs.03062. [DOI] [PubMed] [Google Scholar]
- 140.Zamora R, Vodovotz Y, Billiar TR. Inducible nitric oxide synthase and inflammatory diseases. Mol Med. 2000;6:347–73. [PMC free article] [PubMed] [Google Scholar]
- 141.Reynolds CM, Suliman HB, Hollingsworth JW, Welty-Wolf KE, Carraway MS, Piantadosi CA. Nitric oxide synthase-2 induction optimizes cardiac mitochondrial biogenesis after endotoxemia. Free Radic Biol Med. 2009;46:564–72. doi: 10.1016/j.freeradbiomed.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Deocaris CC, Kaul SC, Wadhwa R. On the brotherhood of the mitochondrial chaperones mortalin and heat shock protein 60. Cell Stress Chaperones. 2006;11:116–28. doi: 10.1379/CSC-144R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Westermann B. Mitochondrial fusion and fission in cell life and death. Nat Rev Mol Cell Biol. 2010;11:872–84. doi: 10.1038/nrm3013. [DOI] [PubMed] [Google Scholar]
- 144.Nakamura T, Lipton SA. Redox regulation of mitochondrial fission, protein misfolding, synaptic damage, and neuronal cell death: potential implications for Alzheimer’s and Parkinson’s diseases. Apoptosis. 2010;15:1354–63. doi: 10.1007/s10495-010-0476-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Cho DH, Nakamura T, Fang J, Cieplak P, Godzik A, Gu Z, Lipton SA. S-nitrosylation of Drp1 mediates beta-amyloid-related mitochondrial fission and neuronal injury. Science. 2009;324:102–5. doi: 10.1126/science.1171091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Gu Z, Nakamura T, Lipton SA. Redox reactions induced by nitrosative stress mediate protein misfolding and mitochondrial dysfunction in neurodegenerative diseases. Mol Neurobiol. 2010;41:55–72. doi: 10.1007/s12035-010-8113-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Cabiscol E, Levine RL. The phosphatase activity of carbonic anhydrase III is reversibly regulated by glutathiolation. Proc Natl Acad Sci U S A. 1996;93:4170–4. doi: 10.1073/pnas.93.9.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Mohr S, Hallak H, de Boitte A, Lapetina EG, Brune B. Nitric oxide-induced S-glutathionylation and inactivation of glyceraldehyde-3-phosphate dehydrogenase. J Biol Chem. 1999;274:9427–30. doi: 10.1074/jbc.274.14.9427. [DOI] [PubMed] [Google Scholar]
- 149.Ji Y, Akerboom TP, Sies H, Thomas JA. S-nitrosylation and S-glutathiolation of protein sulfhydryls by S-nitroso glutathione. Arch Biochem Biophys. 1999;362:67–78. doi: 10.1006/abbi.1998.1013. [DOI] [PubMed] [Google Scholar]
- 150.Lash LH. Mitochondrial glutathione transport: physiological, pathological and toxicological implications. Chem Biol Interact. 2006;163:54–67. doi: 10.1016/j.cbi.2006.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Go YM, Jones DP. Redox compartmentalization in eukaryotic cells. Biochim Biophys Acta. 2008;1780:1273–90. doi: 10.1016/j.bbagen.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Steffen M, Sarkela TM, Gybina AA, Steele TW, Trasseth NJ, Kuehl D, Giulivi C. Metabolism of S-nitrosoglutathione in intact mitochondria. Biochem J. 2001;356:395–402. doi: 10.1042/0264-6021:3560395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Hill BG, Bhatnagar A. Role of glutathiolation in preservation, restoration and regulation of protein function. IUBMB Life. 2007;59:21–6. doi: 10.1080/15216540701196944. [DOI] [PubMed] [Google Scholar]
- 154.Basu S, Keszler A, Azarova NA, Nwanze N, Perlegas A, Shiva S, Broniowska KA, Hogg N, Kim-Shapiro DB. A novel role for cytochrome c: Efficient catalysis of S-nitrosothiol formation. Free Radic Biol Med. 2010;48:255–63. doi: 10.1016/j.freeradbiomed.2009.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Bolanos JP, Heales SJ, Peuchen S, Barker JE, Land JM, Clark JB. Nitric oxide-mediated mitochondrial damage: a potential neuroprotective role for glutathione. Free Radic Biol Med. 1996;21:995–1001. doi: 10.1016/s0891-5849(96)00240-7. [DOI] [PubMed] [Google Scholar]
- 156.Chen T, Pearce LL, Peterson J, Stoyanovsky D, Billiar TR. Glutathione depletion renders rat hepatocytes sensitive to nitric oxide donor-mediated toxicity. Hepatology. 2005;42:598–607. doi: 10.1002/hep.20813. [DOI] [PubMed] [Google Scholar]
- 157.Gegg ME, Beltran B, Salas-Pino S, Bolanos JP, Clark JB, Moncada S, Heales SJ. Differential effect of nitric oxide on glutathione metabolism and mitochondrial function in astrocytes and neurones: implications for neuroprotection/neurodegeneration? J Neurochem. 2003;86:228–37. doi: 10.1046/j.1471-4159.2003.01821.x. [DOI] [PubMed] [Google Scholar]
- 158.Yap LP, Garcia JV, Han DS, Cadenas E. Role of nitric oxide-mediated glutathionylation in neuronal function: potential regulation of energy utilization. Biochem J. 2010;428:85–93. doi: 10.1042/BJ20100164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Garcia J, Han D, Sancheti H, Yap LP, Kaplowitz N, Cadenas E. Regulation of mitochondrial glutathione redox status and protein glutathionylation by respiratory substrates. J Biol Chem. 2010;285:39646–54. doi: 10.1074/jbc.M110.164160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Lundberg M, Johansson C, Chandra J, Enoksson M, Jacobsson G, Ljung J, Johansson M, Holmgren A. Cloning and expression of a novel human glutaredoxin (Grx2) with mitochondrial and nuclear isoforms. J Biol Chem. 2001;276:26269–75. doi: 10.1074/jbc.M011605200. [DOI] [PubMed] [Google Scholar]
- 161.Hashemy SI, Johansson C, Berndt C, Lillig CH, Holmgren A. Oxidation and S-nitrosylation of cysteines in human cytosolic and mitochondrial glutaredoxins: effects on structure and activity. J Biol Chem. 2007;282:14428–36. doi: 10.1074/jbc.M700927200. [DOI] [PubMed] [Google Scholar]
- 162.Lima B, Forrester MT, Hess DT, Stamler JS. S-nitrosylation in cardiovascular signaling. Circ Res. 2010;106:633–46. doi: 10.1161/CIRCRESAHA.109.207381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Liu L, Hausladen A, Zeng M, Que L, Heitman J, Stamler JS. A metabolic enzyme for S-nitrosothiol conserved from bacteria to humans. Nature. 2001;410:490–4. doi: 10.1038/35068596. [DOI] [PubMed] [Google Scholar]
- 164.Holmgren A, Lu J. Thioredoxin and thioredoxin reductase: current research with special reference to human disease. Biochem Biophys Res Commun. 2010;396:120–4. doi: 10.1016/j.bbrc.2010.03.083. [DOI] [PubMed] [Google Scholar]
- 165.Jneid H, Chandra M, Alshaher M, Hornung CA, Tang XL, Leesar M, Bolli R. Delayed preconditioning-mimetic actions of nitroglycerin in patients undergoing exercise tolerance tests. Circulation. 2005;111:2565–71. doi: 10.1161/CIRCULATIONAHA.104.515445. [DOI] [PubMed] [Google Scholar]
- 166.Lin J, Steenbergen C, Murphy E, Sun J. Estrogen receptor-beta activation results in S-nitrosylation of proteins involved in cardioprotection. Circulation. 2009;120:245–54. doi: 10.1161/CIRCULATIONAHA.109.868729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Haendeler J, Hoffmann J, Zeiher AM, Dimmeler S. Antioxidant effects of statins via S-nitrosylation and activation of thioredoxin in endothelial cells: a novel vasculoprotective function of statins. Circulation. 2004;110:856–61. doi: 10.1161/01.CIR.0000138743.09012.93. [DOI] [PubMed] [Google Scholar]
- 168.Bolli R, Bhatti ZA, Tang XL, Qiu Y, Zhang Q, Guo Y, Jadoon AK. Evidence that late preconditioning against myocardial stunning in conscious rabbits is triggered by the generation of nitric oxide. Circ Res. 1997;81:42–52. doi: 10.1161/01.res.81.1.42. [DOI] [PubMed] [Google Scholar]
- 169.Nadtochiy SM, Burwell LS, Ingraham CA, Spencer CM, Friedman AE, Pinkert CA, Brookes PS. In vivo cardioprotection by S-nitroso-2-mercaptopropionyl glycine. J Mol Cell Cardiol. 2009;46:960–8. doi: 10.1016/j.yjmcc.2009.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Bolli R. Cardioprotective function of inducible nitric oxide synthase and role of nitric oxide in myocardial ischemia and preconditioning: an overview of a decade of research. J Mol Cell Cardiol. 2001;33:1897–918. doi: 10.1006/jmcc.2001.1462. [DOI] [PubMed] [Google Scholar]