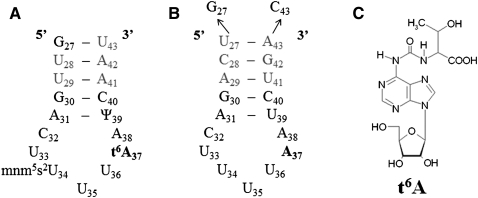Abstract
The post-transcriptional nucleoside modifications of tRNA's anticodon domain form the loop structure and dynamics required for effective and accurate recognition of synonymous codons. The N6-threonylcarbamoyladenosine modification at position 37 (t6A37), 3′-adjacent to the anticodon, of many tRNA species in all organisms ensures the accurate recognition of ANN codons by increasing codon affinity, enhancing ribosome binding, and maintaining the reading frame. However, biosynthesis of this complex modification is only partially understood. The synthesis requires ATP, free threonine, a single carbon source for the carbamoyl, and an enzyme yet to be identified. Recently, the universal protein family Sua5/YciO/YrdC was associated with t6A37 biosynthesis. To further investigate the role of YrdC in t6A37 biosynthesis, the interaction of the Escherichia coli YrdC with a heptadecamer anticodon stem and loop of lysine tRNA (ASLLysUUU) was examined. YrdC bound the unmodified ASLLysUUU with high affinity compared with the t6A37-modified ASLLysUUU (Kd = 0.27 ± 0.20 μM and 1.36 ± 0.39 μM, respectively). YrdC also demonstrated specificity toward the unmodified versus modified anticodon pentamer UUUUA and toward threonine and ATP. The protein did not significantly alter the ASL architecture, nor was it able to base flip A37, as determined by NMR, circular dichroism, and fluorescence of 2-aminopuine at position 37. Thus, current data support the hypothesis that YrdC, with many of the properties of a putative threonylcarbamoyl transferase, most likely functions as a component of a heteromultimeric protein complex for t6A37 biosynthesis.
Keywords: substrate binding, fluorescence, 2-aminopurine, enzyme mechanism, t6A37 synthetase
INTRODUCTION
The high fidelity of translation is critical to the formation of properly functioning proteins. Inaccurate decoding at the ribosome can result in a myriad of diseases and afflictions. Therefore, it is crucial that transfer RNAs (tRNAs) have high affinity and precise recognition of their cognate, synonymous codons. Post-transcriptional modifications of nucleosides in the anticodon stem and loop domain (ASL) are known to play a critical role in maintaining the accuracy of translation (Björk 1995; Agris 1996, 2008). Two of the most frequently modified nucleosides in the ASL are found at the first, or wobble, position 34 of the anticodon and at the universal purine position 37, 3′-adjacent to the anticodon (Nishimura 1972; Agris 1996). Nucleosides 34 and 37 are also two of the most diversely and chemically rich modified nucleosides found in all RNAs (Agris 1996; Jühling et al. 2009). These modifications define anticodon architecture, restrict dynamics of the ASL, enhance codon affinity and ribosome binding, as well as reduce translational frameshifting (Urbonavicius et al. 2001; Agris 2004; Lin et al. 2010).
The highly conserved purine 37 has extensive modification chemistries, each requiring multiple enzymatic steps (Agris 1996). Of these, the universal N6-threonylcarbamoyladenosine modification (t6A37) ensures the accurate recognition of ANN codons (Fig. 1A,C; Nishimura 1972). The t6A37-modified nucleoside has been shown to be essential for ribosome-mediated codon binding by tRNALysUUU, tRNAArgUCU, and tRNAIleGAU and stabilizes anticodon interaction with the cognate and wobble codons on the ribosome-bound mRNA (Miller et al. 1976; Weissenbach and Grosjean 1981; Yarian et al. 2002). This modification has also been shown to be an identity determinant for recognition of the cognate tRNA by at least one aminoacyl-tRNA synthetase (Nureki et al. 1994). Structural analysis by nuclear magnetic resonance spectroscopy (NMR) and X-ray crystallography has shown that the t6A37 modification of tRNALysUUU is required for proper anticodon architecture to induce codon binding (Stuart et al. 2000; Murphy et al. 2004). The modification creates an open, but ordered and structured, architecture for the loop by negating intraloop base-pairing of U33•A37 and by enhanced base stacking on the 3′ side of the anticodon (Stuart et al. 2000).
FIGURE 1.
(A) Nucleotide sequence of the fully modified E. coli tRNALysUUU anticodon stem and loop domain. (B) Sequence of the unmodified human tRNALysUUU anticodon stem and loop domain, ASLLysUUU. The terminal U27•A43 base pair of the natural sequence has been replaced by a G27•C43 base pair to increase the stability of the stem. (C) The N6-threonylcarbomoyladenosine (t6A37) modification found at position 37 of tRNALysUUU.
Although this modification is required for accurate translation, the mechanism of t6A37 biosynthesis is not well understood. The reaction requires ATP, free threonine, and a single carbon source (Chheda et al. 1972; Powers and Peterkofsky 1972; Elkins and Keller 1974; Körner and Söll 1974). Studies of t6A biosynthesis in vitro have demonstrated that Escherichia coli extracts contain an enzymatic component required for synthesis, but the identity of that enzyme remains unknown (Elkins and Keller 1974; Körner and Söll 1974). Recently, molecular genetic analyses have linked the Sua5/YciO/YrdC protein family with t6A37 formation through mutation studies (El Yacoubi et al. 2009).
YrdC is a member of the universal Sua5/YciO/YrdC family of proteins that is present as separate proteins or domains within larger proteins, and is highly conserved from E. coli to humans. Because of its ubiquitous expression, structure, and homologs' function, YrdC has been thought to play a role in protein synthesis (Chen et al. 2003). E. coli YrdC has also been linked to ribosome biogenesis, specifically 16S rRNA maturation (Kaczanowska and Rydén-Aulin 2004, 2005). However, precise details as to its roles in these functions remain unclear.
The crystal structure of E. coli YrdC, solved as a dimer, revealed an α/β twisted open-sheet with a positively charged concave surface that is suggestive of a double-stranded RNA binding site (Teplova et al. 2000). tRNA binding was experimentally verified and showed YrdC selectively binds tRNAThrCGU lacking the t6A37 modification over its modified counterpart (El Yacoubi et al. 2009). YrdC is essential in E. coli and yrdC expression is required for the formation of t6A37 in E. coli tRNA (Baba et al. 2006; El Yacoubi et al. 2009). The E. coli protein has been shown to preferentially bind ATP and ADP over UTP and CTP; however, ATPase activity required for t6A37 biosynthesis has yet to be observed (El Yacoubi et al. 2009).
The yeast homolog, Sua5, contains a YrdC domain and has been suggested to function in translational regulation (Na et al. 1992). More recently, the gene product of sua5 from Saccharomyces cerevisiae has been shown to play a role in t6A37 biosynthesis (El Yacoubi et al. 2009). In this study sua5 deletion mutants led to the loss of t6A37. Additionally, reading frame maintenance of translation was adversely affected by loss of Sua5 and t6A37 (Lin et al. 2010).
Several co-crystal structures of RNA-modifying enzymes and their substrate RNAs have been solved, providing information about their mechanisms of action (for review, see Byrne et al. 2009). For example, the MnmA protein of E. coli is required for the substitution of a 2-thionyl for the 2-carbonyl of the wobble position uridine-34 in tRNAGlnUUG, tRNAGluUUC, and tRNALysUUU. The co-crystal structure of the MnmA and tRNAGluUUC reveals an interaction of the protein with the tRNA's D-stem and ASL. The U34 is flipped out of the anticodon base stack in order for MnmA to gain access to its substrate (Numata et al. 2006). This interaction appears to be moderated by a base-specific recognition of the anticodon (Kambampati and Lauhon 2003). Another RNA modification enzyme, E. coli MiaA, catalyzes the transfer of the isopentyl-moiety from dimethylallyl pyrophosphate to A37 of tRNAs with an A36-A37-A38 sequence to form i6A37 and has been co-crystalized with the E. coli tRNAPheGAA (Chimnaronk et al. 2009; Seif and Hallberg 2009). The interaction of MiaA with tRNAPheGAA displays a mutually induced fit in which the anticodon loop unfolds, and U33, G34, A35, and A37 are flipped outside of the canonical 3′-base stack of the loop (Seif and Hallberg 2009). This base flipping mechanism makes target nucleosides more accessible and is common to many RNA and DNA modification enzymes (for review, see Roberts and Cheng 1998).
In this study, we combined a series of biochemical and structural approaches to investigate the interactions of E. coli YrdC with its potential substrates. We show that YrdC selectively binds the unmodified ASLLysUUU and threonine. The interaction of YrdC with ASLLysUUU did not appear to induce a structural change in A37, indicating that the enzymatic mechanism for modification does not include base flipping or that YrdC may function in concert with other proteins in order to constitute the enzymatic activity required for t6A37 biosynthesis.
RESULTS
YrdC selectively binds unmodified ASLLysUUU
The tRNAs responding to ANN codons and having the t6A37 modification or a derivative of it such as m6t6A37 or ms2t6A37, depending on tRNA species and organism, include tRNAs for arginine, isoleucine, lysine, and threonine. The affinity of YrdC for the full-length E. coli tRNAThrCGU lacking only the t6A37 modification has been shown through determination of equilibrium dissociation constants (Kd = 0.11 ± 0.05 μM) to be greater than that of its fully modified counterpart (Kd = 0.62 ± 0.13 μM) (El Yacoubi et al. 2009). Because the site of threonylcarbamoylation is within the anticodon domain of the tRNA, we wanted to determine if the heptadecamer ASL (Fig. 1) was sufficient for recognition and binding by YrdC. Change in the intrinsic fluorescence of the two tryptophans of YrdC (residues W89 and W106) was utilized to measure the protein's binding and to calculate apparent binding constants for variously modified ASLs, smaller oligonucleosides, mononucleoside triphosphates, and amino acids. First, YrdC was titrated with several unmodified ASLs to determine its ability to recognize specifically the ASL of an unmodified tRNA substrate. The anticodon stem and loop sequence of the human tRNALysUUU (ASLLysUUU) (Fig. 1B) was used because the base pair content of its stem contributes considerably more stability to this small hairpin than that of the E. coli ASLLysUUU. The stem of the human ASLLysUUU is stabilized by two internal G•C base pairs and a terminal G•C base pair substituting for an A•U in order to conduct NMR and crystallographic studies (Fig. 1B). Human ASLLysUUU and the E. coli ASLLysUUU have otherwise identical anticodon loop sequences and closely related modified nucleosides (Jühling et al. 2009). Previous studies have successfully used the human ASLLysUUU as a common context to study the structural effects of both the E. coli and human modifications (Durant et al. 2005).
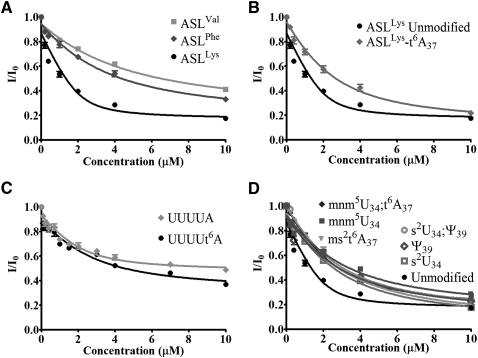
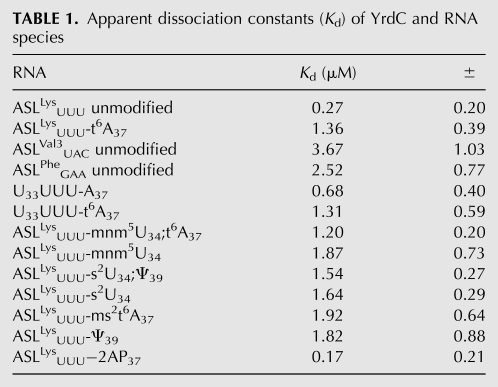
The tryptophan fluorescence of YrdC was quenched by almost 80% with the addition of increasing concentrations of ASLLysUUU and at an ASLLysUUU/protein ratio of as little as 2/1. E. coli ASLVal3UAC has an A37, but it is modified to m6A37. Yeast ASLPheGAA has a G37, and it is modified to wyobutosine (yW37). In contrast to the titration of YrdC with ASLLysUUU, its titration with E. coli ASLVal3UAC or yeast ASLPheGAA resulted in at most a 60% reduction in fluorescence at a ratio of 5/1. The affinity of YrdC for unmodified ASLLysUUU, as determined from the equilibrium dissociation constant (Kd = 0.27 ± 0.2 μM), was significantly greater than that of the other unmodified ASLs that had been tested (Fig. 2A; Table 1). Thus, the recognition of ASLLysUUU by YrdC in comparison to that for ASLVal3UAC and ASLPheGAA implies some sequence-dependent specificity.
FIGURE 2.
Changes in YrdC fluorescence with the addition of differentially modified ASLs. (A) The titration of YrdC with unmodified ASLLysUUU, ASLVal3UAC, and yeast ASLPheGAA. (B) The titration of YrdC with the unmodified ASLLysUUU and the t6A37-modified ASLLysUUU-t6A37. (C) The titration of YrdC with nucleoside pentamers analogous to the tRNALys anticodon region, U33UUUA37, and U33UUUt6A37. (D) The titration of YrdC with variously modified ASLLysUUU with the modifications 5-methylaminomethyluridine, mnm5U34, 2-thiouridine, s2U34, pseudouridine, Ψ39, and the 2-methylthio derivative of t6A37, ms2t6A37.
TABLE 1.
Apparent dissociation constants (Kd) of YrdC and RNA species
Should YrdC be the threonylcarbamoyltransferase or a subunit of the enzyme, it would be expected to have a lower affinity for the product of the reaction, the modified ASLLysUUU-t6A37, than for the substrate, the unmodified ASLLysUUU. We assessed the affinity of YrdC for the ASLLysUUU-t6A37 and found that YrdC bound the ASLLysUUU-t6A37 with a significantly lower affinity, Kd = 1.36 ± 0.39 μM (Fig. 2B; Table 1). This preferred recognition of the unmodified ASL substrate, taken together with the specificity for the ASLLysUUU, suggests that YrdC may very well be involved in t6A37 synthesis through its direct interaction with the ASL, specifically A37. These results are consistent with the previously reported binding of YrdC to full-length tRNAThrCGU where the only modification missing from the tRNAThrCGU was the t6A37. The difference in binding affinity of YrdC to the full-length, fully modified tRNAThrCGU versus the tRNAThrCGU lacking only the t6A modification was approximately fivefold (El Yacoubi et al. 2009), comparable to the differences in affinity we have observed for ASLLysUUU-t6A37 and unmodified ASLLysUUU.
To determine if YrdC would recognize a tRNALysUUU fragment as small as a pentamer and to further test its specificity for unmodified A37, the protein's ability to bind and discriminate between two anticodon region oligonucleotides was examined (Fig. 2C). The oligomers U33UUUA37 and U33UUUt6A37 are composed of the invariant U33 responsible for tRNA's canonical “U-turn” just prior to the anticodon, the anticodon UUU, and the substrate nucleoside A37. As evidenced by the dissociation constants derived from the change in tryptophan fluorescence, YrdC exhibited a considerable affinity for the unmodified pentamer in comparison to that for the t6A-modified pentamer (Table 1). The pentamers were not recognized as well as the heptadecamer ASL. The affinity of YrdC for the unmodified pentamer was only a third that for the hairpin ASLLysUUU. It appears that YrdC is able to differentially recognize the anticodon sequence of ASLLysUUU in the absence of the hairpin, stem, and loop structure and to discern between the putative substrate A37 and possible product t6A37.
The affinity of YrdC for differentially modified ASLLysUUU constructs was tested by fluorescence quenching of YrdC to observe the effect that modifications, in general, may have on its recognition of the ASL (Fig. 2D). The completely modified, naturally occurring E. coli tRNALysUUU contains 5-methylaminomethyl-2-thiouridine-34, mnm5s2U34, t6A37 and pseudo-uridine 39, Ψ39 (Fig. 1A). The affinity of YrdC for the unmodified ASLLysUUU was at least fourfold greater than that for the modified ASLLysUUU constructs (Table 1). The ASLLysUUU-mnm5U34;t6A37 and the ASLLysUUU-ms2t6A37 with the mammalian 2-methylthio-t6A37 would be expected to bind with an affinity comparable to that of ASLLysUUU-t6A37 because the A37 in each is modified with an N6-threonylcarbamoyl-moiety. Indeed, these two modified ASLLysUUU bound to YrdC with apparent dissociation constants more reflective of the t6A37-modified ASL (Kd = 1.20 ± 0.20 and 1.92 ± 0.64, respectively) than that of the unmodified ASLLysUUU (Table 1). The modified ASLs lacking t6A37, ASLLysUUU-s2U34, ASLLysUUU-s2U34;Ψ39, ASLLysUUU-Ψ39, and ASLLysUUU-mnm5U34 would be expected to be recognized by YrdC with Kd values similar to the unmodified ASLLysUUU, because they are much closer to the putative natural substrate of t6A37 biosynthesis. Surprisingly, the dissociation constants for the ASLs with s2U34, mnm5U34, and Ψ39 were similar to those for ASLs with the threonylcarbamoyl modification at A37. s2U34, mnm5U34, and mnm5s2U34 are known to contribute order to the ASLLysUUU structure (Stuart et al. 2000; Sundaram et al. 2000; Murphy et al. 2004; Durant et al. 2005). Ψ39, located two nucleosides 3′ to A37, is known to stabilize the 3′ side of the anticodon loop (Durant and Davis 1999; Yarian et al. 1999) Therefore, the contributions of these modifications to the conformation and dynamics of the ASL could impart a structure to the anticodon loop less favorable for YrdC recognition. The chronology of events leading to the fully modified anticodon domain of the tRNA is not known, so it is possible that these post-transcriptional modifications are sequential and that formation of t6A37 is the primary event.
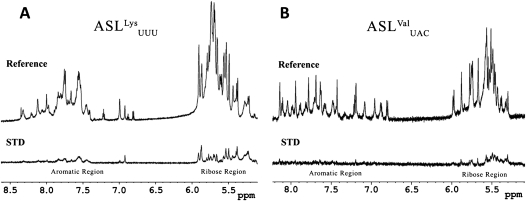
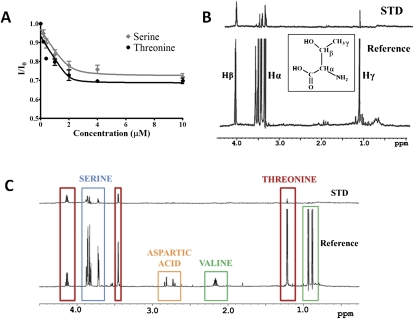
Saturation transfer difference NMR (STD-NMR) was conducted as an additional independent method to verify the unmodified ASLLysUUU binding by YrdC. STD-NMR is able to detect the presence of binding equilibria and provides information to identify protons of ligands in direct contact with the protein (Mayer and Meyer 1999). In the STD spectrum, only the protons representing molecules interacting with the protein will be observable. The reference spectrum depicts the ribose (6–4.5 ppm) and the aromatic (8.5–6.5 ppm) regions for unmodified ASLLysUUU (Fig. 3A) and unmodified ASLVal3UAC (Fig. 3B). In the lower STD spectrum of the unmodified ASLLysUUU, there is considerable recovery of many ribose peaks. There is also a significant recovery of specific resonances in the aromatic region of the spectrum, indicating that YrdC is making contacts with what appears to be a select set of the bases. In contrast, the recovery of the ribose protons of unmodified ASLVal3UAC is less and the aromatic protons show no observable signal. This recognition of the backbone is not unexpected considering the Kd of YrdC for ASLVal3UAC (Table 1). Given these results with the fluorescence quenching data, it is very likely that YrdC recognizes the RNA substrate in a sequence-dependent manner by directly interacting with certain bases.
FIGURE 3.
Saturation transfer difference (STD)-NMR of YrdC with unmodified ASLLysUUU (A) and unmodified ASLVal3UAC (B). For each set of experiments, there is a one-dimensional NMR reference spectrum (top) and a STD spectrum of recovered signals (bottom). The spectral region shown displays the characteristic RNA chemical shifts for the aromatic protons (8.5 to 6.5 ppm) and the ribose protons (6.0 to 5.0 ppm). The STD-NMR spectrum records the proton signals from those residues of the ASL that have been bound by YrdC. The standard one-dimensional 1H-NMR recapitulate previously published data for the unmodified ASLLysUUU (Stuart et al. 2000) and unmodified ASLVal3UAC (Vendeix et al. 2008).
YrdC recognizes threonine and ATP
The biosynthesis of t6A37 utilizes free threonine from the cell as the threonyl-moiety for production of the threonylcarbamoyl (Powers and Peterkofsky 1972). If YrdC is involved in the enzymatic pathway, it could potentially bind threonine, ATP, and/or the carboxylphosphate. The binding of threonine by YrdC was determined by monitoring the protein's tryptophan fluorescence while titrating with the amino acid (Fig. 4A). YrdC bound threonine with a Kd = 0.04 ± 0.10 μM. This very tight binding indicates the likelihood of a specific binding site on YrdC for threonine. The same experiment was conducted with serine. The binding affinity of YrdC for threonine was threefold higher than that for serine (Kd = 0.12 ± 0.16 μM).
FIGURE 4.
Changes in fluorescence of YrdC with additions of the amino acids threonine and serine (A) and STD-NMR of threonine in the presence of YrdC (B). The one-dimensional reference spectrum with 1 mM threonine (bottom) and the STD spectrum (top). (C) Competition of amino acids in binding to YrdC. The STD-NMR experiment was conducted with YrdC and equal molar concentrations of serine (blue), threonine (red), aspartic acid (orange), and valine (green).
STD-NMR was also conducted to identify the binding epitope of threonine. In the STD spectrum, only the peaks representing bound protons will be seen. A STD signal was observed with 1 mM threonine, and all of the peaks were recovered (Fig. 4B). It appears from the differential changes in the threonine resonances that YrdC binds threonine with a preference for the Hβ. An additional STD-NMR experiment was performed to observe the selectivity of YrdC with several amino acids. Because of their similarities, serine, valine, and aspartic acid were added together with threonine to compete for binding to YrdC. It appears that YrdC can in fact preferentially bind threonine over the other amino acids present (Fig. 4C). YrdC also recognized serine, but to a lesser extent than threonine, as determined by integration of the peaks. The peak at 4.1 ppm represents the Hβ of threonine and is recovered by 48%. The Hα threonine peak is 14% recovered, and the serine peaks were only 10% and 8% recovered, respectively. This is most likely due to the recognition of serine's and threonine's common elements. Since valine is not bound, we believe YrdC must be distinguishing the OH that serine and threonine have in common, and not the methyl group.
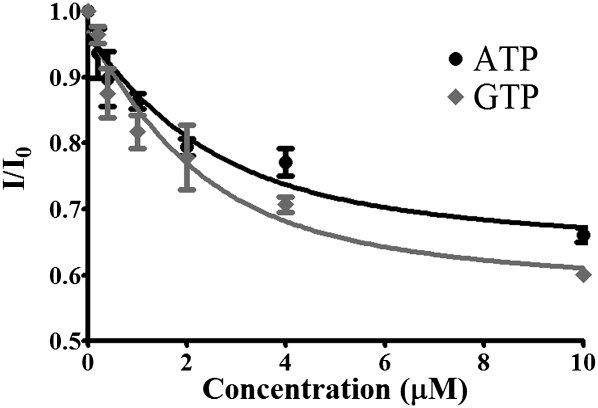
YrdC's preferential binding of ATP and ADP had been reported previously; however, the protein's affinity for the nucleotides had not been determined (El Yacoubi et al. 2009). Using tryptophan fluorescence quenching, YrdC was titrated with ATP and GTP (Fig. 5). The protein bound ATP and GTP with estimated Kd values of 1.04 ± 0.67 μM and 1.31 ± 0.91 μM, respectively.
FIGURE 5.
YrdC binding of ATP and GTP. YrdC was titrated with ATP and GTP and binding of the nucleotide-5′-triphosphates was measured by observing changes in tryptophan fluorescence.
YrdC does not induce a significant change in ASLLysUUU structure, particularly at A37
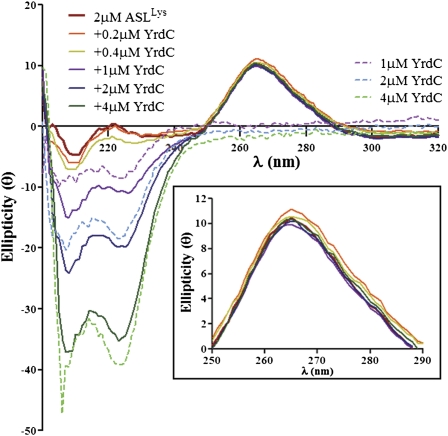
Modification of A37 to t6A37 may involve a conformational perturbation of the anticodon stem and loop or, minimally, a conformational change in A37. Evidently, a conformational change in the tRNA is not required for all modification recognition mechanisms. The protein MnmA, responsible for thiolation of U34 recognizes tRNA in a predominantly rigid body manner by flipping out U34 without affecting the rest of the tRNA structure (Numata et al. 2006). However, all other co-crystals of RNA-modifying enzymes with their tRNA substrates to date have depicted structural changes in the tRNA and/or the enzyme (for reviews, see Ishitani et al. 2008; Byrne et al. 2009). For that reason, we employed circular dichroism (CD) spectroscopy to determine if YrdC induces a conformational change in the unmodified ASLLysUUU. CD spectra are sensitive to structural changes in base stacking within an RNA and in response to the binding of a protein, changes in temperature, pH, and metal binding (Chen et al. 1993; Agris et al. 1999). Changes in base stacking are monitored by the positive ellipticity found between 250 and 290 nm. This spectral window is particularly advantageous for observing the RNA in presence of YrdC because proteins have a spectral null in this exact wavelength range. ASLLysUUU was titrated with increasing concentrations of YrdC to observe the effect the protein had on the RNA structure (Fig. 6). Each spectrum representing a different concentration of YrdC was within experimental error of the spectrum in the absence of the protein and therefore, reported no global change in base stacking. Even at twice the concentration of the RNA, YrdC did not induce an observable change in the ASL.
FIGURE 6.
Circular dichroism (CD) spectra of the unmodified ASLLysUUU titrated with YrdC. The unmodified ASLLysUUU (2 μM) was titrated with YrdC (0.2–4.0 μM), and the resulting CD spectra were collected. In addition, the spectra of YrdC alone were collected at the same concentrations. A magnification of the 250- to 290-nm spectral region is shown to examine changes in the RNA base stacking with addition of YrdC (inset).
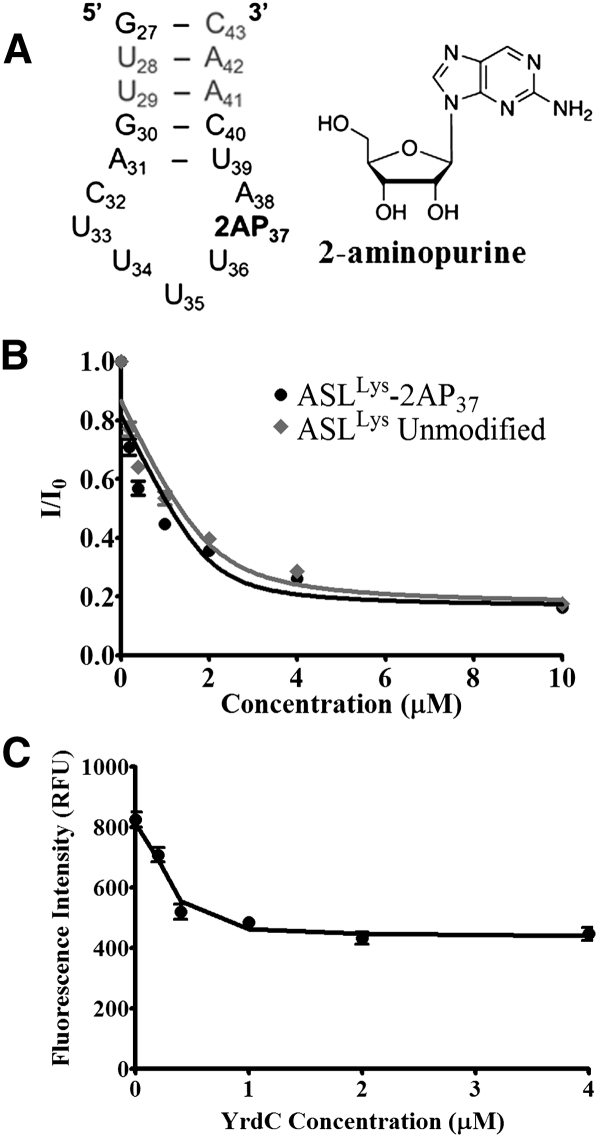
An alteration of only the A37 nucleoside conformation may not have been observable by CD because the nucleoside was one of 17 residues being averaged in the spectroscopy. Therefore, to investigate any possible perturbation of the A37 with the binding of YrdC, ASLLysUUU was synthesized with a fluorescent analog of adenosine, 2-aminopurine ribonucleoside, at position 37 (2AP37) (Fig. 7A). Though not capable of a Watson-Crick base-pairing, 2AP in loops is commonly used to probe nucleic acid structure because it rarely affects structure adversely, and its fluorescence is typically enhanced when its environment is disturbed as a result of decreased stacking interactions (Rachofsky et al. 2001). Therefore, if a conformational change of A37 occurs such as flipping outside of the anticodon loop base stack, an increase in the fluorescence would be expected. To ensure that YrdC recognition of the ASLLysUUU-2AP37 was comparable to that of the unmodified ASLLysUUU, the affinity was assessed by monitoring tryptophan fluorescence as described above (Fig. 7B). YrdC recognized ASLLysUUU-2AP37 with a Kd = 0.17 ± 0.21 μM, comparable to that for ASLLysUUU. The 2AP fluorescence of ASLLysUUU-2AP37 (λex = 310nm, λem = 375 nm) was monitored while titrating the ASLLysUUU with YrdC. With the addition of YrdC, the 2AP fluorescence is quenched (Fig. 7C). This suggests that upon protein binding, A37 is stacking with either adjacent bases or aromatic amino acids and is not being flipped to a position exposed to solvent. Since 2AP37 is within the flexible loop region, it is likely that binding of YrdC creates a more structured local environment for A37, possibly in the binding pocket.
FIGURE 7.
Nucleoside A37 monitored for base flipping. (A) The nucleoside sequence of the 2-aminopurine ribonucleoside (2AP37) construct. (B) Fluorescence changes in the tryptophans of YrdC with addition of the unmodified ASLLysUUU and the 2AP37-modified ASLLysUUU-2AP37. (C) Changes in fluorescence of 2AP37 (λex = 310 nm; λem = 375 nm) with addition of increasing concentrations of YrdC.
Two physicochemical approaches, CD and 2AP fluorescence, indicated that the structure and conformational dynamics of the ASLLysUUU, and those of the target A37, were not significantly altered when bound by YrdC. In order to determine at the level of atomic resolution whether or not the ASL or the nucleoside were subjected to a structural change, we employed NMR spectroscopy. The nucleoside resonances for ASLLysUUU had been assigned previously (Durant and Davis 1999; Stuart et al. 2000), and the chemical shifts of many of the aromatic resonances of the RNA, including cross-peaks for A37, were clearly observable in the presence of the protein (Fig. 8). After collecting a Nuclear Overhauser Effect spectrum (NOESY) of the ASLLysUUU alone, it was titrated with YrdC to a final stoichiometry of 1:1 and another spectrum collected. No significant changes occurred in the resonances of the RNA when observing the nucleobase aromatic protons. However, the other substrates required for the biosynthesis of t6A37, ATP, threonine, and what is suspected to be an activated bicarbonate such as carboxylphosphate, were missing from the experiment. Therefore, ATP and then threonine were added to the YrdC-ASLLysUUU sample in two steps. Again, the NMR spectra depicted no significant peak movement or broadening in the ASL amino region. All four of the spectra overlay completely with minor shifts in some resonances for A37 and A38 and slight shifts in the loop uridines. With the addition of threonine, line broadening was observed and resulted in the loss of several peaks (data not shown). This could be due to a number of reasons including the YrdC binding of the RNA or the high concentration of molecules in the sample decreasing tumbling.
FIGURE 8.
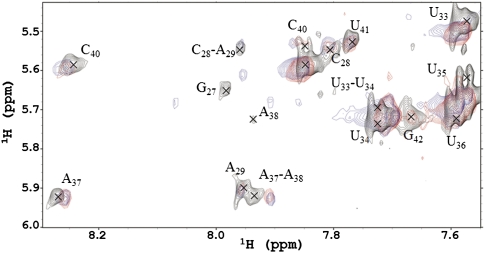
1H-NMR NOESY spectra of unmodified ASLLysUUU with additions of YrdC and ATP. The spectra were collected in 10% D2O at 25°C. A region of the ASLLysUUU spectrum (7.4–8.4 ppm × 5.4–6.0 ppm) is practically free of overlapping resonances from the protein. The connectivities between the ASLLysUUU aromatic protons and the H1′ of the ribose are annotated: black, unmodified ASLLysUUU alone; blue, addition of YrdC; and red, addition of ATP to YrdC and the ASL.
DISCUSSION
In 1974, an enzymatic activity capable of the synthesis of t6A in a t6A-deficient tRNA was found in a crude extract from an E. coli culture (Elkins and Keller 1974; Körner and Söll 1974). Over 30 years later, there is little information about the enzyme(s) responsible for this reaction. Molecular genetic analyses point to the Sua5/YciO/YrdC protein family as having a role in t6A37 formation. The knockdown of the yrdC gene results in a loss of t6A37 in E. coli tRNAs, and the knockout of the sua5 gene in S. cerevisiae causes inadequate t6A37 production (El Yacoubi et al. 2009; Lin et al. 2010). Because of its double-stranded RNA-binding ability and suspected ATPase activity, YrdC has emerged as the most likely candidate for the role of N6-adenosine threonylcarbamoyltransferase in E. coli.
YrdC is able to recognize small hairpin RNAs, more specifically the ASL of tRNALysUUU, as we have shown by tryptophan fluorescence quenching, STD-NMR, and novel Raman spectroscopy techniques (data not shown). It binds with sequence specificity and preferentially binds unmodified ASLLysUUU compared with the modified counterpart, ASLLysUUU-t6A37, and the unmodified ASLVal3UAC and yeast ASLPheGAA. In fact, YrdC binds the unmodified ASLLysUUU with an affinity (Kd = 0.27 ± 0.20 μM) very comparable to its reported recognition of the full-length, native E. coli tRNAThrCGU, lacking only the t6A37 (Kd = 0.11 ± 0.05 μM) (El Yacoubi et al. 2009). YrdC binding of the ASLLysUUU-t6A37 exhibited a fivefold increase in Kd compared with that for the unmodified ASLLysUUU. This result is also analogous to the previously reported full-length, fully modified tRNAThrCGU versus tRNAThrCGU lacking only t6A37 (El Yacoubi et al. 2009). YrdC exhibited an affinity for ASLLysUUU similar to that for the full-length tRNAThrCGU lacking t6A37, indicating that most of the important binding determinants are within the ASL region of the tRNA and not within other domains. The twofold difference in Kd between the respective pairs (fully modified tRNAThrCGU versus the ASLLysUUU-t6A, and the tRNAThrCGU lacking only t6A37 versus the unmodified ASLLysUUU) could be the result of YrdC interactions with more than just the anticodon domain or could reflect differences in recognition of the two natural substrate ASL sequences (tRNAThrCGU vs. tRNALysUUU). At this time, our results do not distinguish between these two explanations for the difference in equilibrium binding constant. The previously reported lack of binding to a fully unmodified tRNAThrCGU transcript (El Yacoubi et al. 2009) could be due to the importance of other modifications outside of the anticodon domain or to the tertiary folding of the tRNA (Helm 2006) and not due to a requirement for a direct interaction with YrdC.
Many modification enzymes display a mechanism of action that introduces a structural change in the tRNA to create a more accessible nucleoside for modification (Ishitani et al. 2008; Byrne et al. 2009). Typically the nucleobase to be modified is flipped, removing it from an internal stacking interaction and perhaps hydrogen bonding, and exposes it to the solvent and the protein active site. Unmodified A37 is located within the anticodon loop of ASLLysUUU (Durant and Davis 1999; Stuart et al. 2000). Therefore, we hypothesized that a base flip would be required for the enzymatic addition of the threonylcarbamoyl-group to form t6A37. However, our present studies do not indicate a base flipping mechanism. Results from titrating of the unmodified ASLLysUUU with YrdC and monitoring either the CD spectra or the fluorescence of 2AP37 did not indicate that a change in structure of the ASL had taken place. This may be due to the absence of other protein components required for catalysis, a requirement for the hydrolysis of ATP, or the limitations of these two methods in exhibiting a change. NMR spectra of the unmodified ASLLysUUU in the presence of YrdC followed by additions of ATP and threonine also did not show a noticeable structural change to the ASL or movement of A37. It is possible that the NMR solution conditions may not have been optimal for observation of a conformational change, as in vitro t6A37 biosynthesis has been shown to require magnesium and bicarbonate (Elkins and Keller 1974).
The proposed t6A37 biosynthesis requires two ATP-dependent steps: formation of carboxylphosphate from bicarbonate and ATP and its transfer to the amino group of adenosine to form N6-carbamoyladenosine, and then activation of the carbamoyl moiety of N6-carbamoyladenosine with a second ATP followed by addition of threonine via attack by its α-amine (Garcia and Goodenough-Lasua 1998). It is possible that YrdC is involved in the latter step of the pathway by positioning threonine and the tRNA substrate into the active site of the protein responsible for catalytic activity. Further research is necessary to elucidate this enzymatic biosynthesis. However, the data presented here along with prior research support the hypothesis that YrdC is a subunit of a complex that constitutes the enzymatic properties for t6A37 biosynthesis where YrdC selectively recognizes and binds the tRNA substrates, ATP and threonine, and the holoenzyme functions as the threonylcarbamoyl transferase. Another possible scenario, based on the binding affinities alone and not on cell concentrations of the ligands, has YrdC binding tRNA and threonine, but not ATP. The observed ATP binding may be nonspecific due to recognition of ATP in the A37 binding pocket on the protein. An additional subunit of the enzyme complex may have the ATPase activity. Recently, the KEOPS/EKC complex in yeast has been shown to be required for t6A37 biosynthesis (Srinivasan et al. 2011). A member of this complex, Kae1, is an ATPase and is highly conserved. Its bacterial ortholog, YgjD, was also shown to be required for modification in E. coli. Further examination of the structural and biochemical interactions between YrdC, its substrates, and the other possible subunits of the threonylcarbamoyl transferase should offer more information about this essential enzymatic pathway.
MATERIALS AND METHODS
Sample preparation and purification
All ASL samples except one were chemically synthesized by Dharmacon (Thermo Fisher) using “ACE” chemistry (Scaringe et al. 1998). The ASLs were de-protected per manufacturer's instructions and dialyzed against 25 mM potassium phosphate buffer (pH 6.8), and 100 mM NaCl using a 3500-Da cutoff. The ASLLysUUU-t6A37 was chemically synthesized with the 2′,5′-protected 3′-phosphoramidite of t6A37 by the Nucleic Acid Facility of North Carolina State University. The oligonucleoside pentamers UUUUA and UUUUt6A were chemically synthesized in solution using the phosphotriester chemistry (Agris et al. 1997).
His6-tagged E. coli YrdC protein was overexpressed in E. coli as described previously and purified by Ni-NTA affinity chromatography to a purity of >95% by SDS-PAGE (El Yacoubi et al. 2009). Protein integrity was verified by crystallography.
Fluorescence quenching
The inherent fluorescence of the two tryptophans present in YrdC was observed at room temperature using a SpectraMax Gemini XS microplate reader (Molecular Devices). Data were collected using accompanying SoftMax Pro software (Molecular Devices). For fluorescence quenching of YrdC, the tryptophans were excited at 295 nm and their emission collected at 345 nm, similar to the method described by Teplova et al. (2000). Triplicate samples of the RNAs, NTPs, and amino acids were lyophilized to dryness in amounts corresponding to the required final concentrations. The titration was performed by resuspending lyophilized samples in 350 μL of 2 μM YrdC (25 mM potassium phosphate, 100 mM NaCl at pH 6.8) to final concentrations of 0.2–10 μM in 96-well microplates. Data were averaged and fit to a single site nonlinear regression,
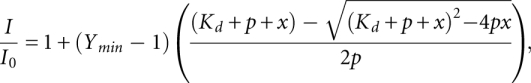 |
where p represents the protein concentration and x equals the ligand concentration. Apparent dissociation constants were calculated with standard errors using this equation according to method described by Kurganov et al. (1972) (GraphPad Prism 5). Error bars represent the standard error of the mean.
The fluorescence of the 2-aminopurine ribonucleoside at position 37 of the ASLLysUUU was excited at 310 nm and the emission collected at 375 nm. YrdC samples were lyophilized to dryness and resuspended to final concentrations of 0.2–10 μM in 350 μL of 2 μM ASLLysUUU-2AP37 sequentially.
CD spectroscopy
CD spectra (200–320 nm) were collected using a Jasco J600 spectropolarimeter at 25°C with a quartz cuvette having 1-cm path length. ASL sample concentrations were adjusted to ∼0.27 A260 U/mL (2 μM). ASL (2 μM in 25 mM potassium phosphate, 100 mM NaCl at pH 6.8) was added to lyophilized YrdC to constitute final protein concentrations of 0.2–10 μM in 1 mL. Each experiment was performed in triplicate. The spectra of YrdC alone at the same concentrations were collected and overlayed. Data were plotted using Excel 2007 (Microsoft) and GraphPad Prism 5.
NMR spectroscopy
STD-NMR data were acquired on a Bruker Avance II 700 MHz spectrometer at UAlbany equipped with an ultra-sensitive triple resonance cryoprobe capable of applying pulsed field gradients along the z-axis. All experiments were conducted at 25°C. For the experiments with RNA, 3 μM YrdC and 300 μM ASLLysUUU or 200 μM ASLVal3UAC was prepared in 300 μL of 99% D2O (10 mM potassium phosphate, 100 mM NaCl at pH 6.8). The STD pulse was applied according the method described by Mayer and Meyer (1999), except the on-resonance irradiation was centered at 0 ppm in order to selectively saturate only protein. A total of 1028 scans with 16 dummy scans was collected. Amino acid samples were prepared in 600 μL of 99% D2O (10 mM potassium phosphate, 100 mM NaCl at pH 6.8). For each experiment, the concentrations were 20 μM YrdC and 1 mM of each amino acid. The STD pulse sequence was applied according the method described by Mayer and Meyer (1999). A total of 256 scans with 16 dummy scans were collected for each.
NOESY spectra were collected on a DMX Bruker 500 MHz instrument equipped with triple-resonance 1H, 13C, and 15N probe and three-axis pulsed field gradient capabilities at NCSU. All NMR data were processed using NMRPipe (Delaglio et al. 1995). Spectra were analyzed using SPARKY software (Goddard and Kneller 2008). Each 2D NOESY experiment was recorded at 25°C with a 200-msec mixing time using the WATERGATE suppression sequence. A spectrum was collected of ASLLysUUU sample alone (∼1 mM; 25 mM potassium phosphate, 100 mM NaCl at pH 6.8 in 10% D2O). For the additions of 0.4 mM YrdC, 1 mM ATP, and 1 mM threonine, the ASLLysUUU concentration was 0.4 mM in the same buffer conditions and 10% D2O.
ACKNOWLEDGMENTS
We thank Caren J. Stark for critical reading and editing of the manuscript, Dirk Iwata-Reuyl and Valérie de Crécy-Lagard for insightful discussions, and Alex Shekhtman and Swamy Puttamadappa for NMR assistance at UAlbany. We also thank Andrzej Malkiewicz for the two oligonucleotide pentamers and the protected phosphoramidite of t6A that was incorporated into ASLLysUUU. This work was supported by grants from the National Institutes of Health (2-RO1-GM23037), the National Science Foundation (MCB-0548602) to P.F.A. and a gift from Beckman-Coulter, Inc. to M.A.S.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2592411.
REFERENCES
- Agris PF 1996. The importance of being modified: roles of modified nucleosides and Mg2+ in RNA structure and function. Prog Nucleic Acid Res Mol Biol 53: 79–129 [DOI] [PubMed] [Google Scholar]
- Agris PF 2004. Decoding the genome: a modified view. Nucleic Acids Res 32: 223–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agris PF 2008. Bringing order to translation: the contributions of transfer RNA anticodon-domain modifications. EMBO Rep 9: 629–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agris PF, Guenther R, Ingram PC, Basti MM, Stuart JW, Sochacka E, Malkiewicz A 1997. Unconventional structure of tRNALysSUU anticodon explains tRNA's role in bacterial and mammalian ribosomal frameshifting and primer selection by HIV-1. RNA 3: 420–428 [PMC free article] [PubMed] [Google Scholar]
- Agris PF, Marchbank MT, Newman W, Guenther R, Ingram P, Swallow J, Mucha P, Szyk A, Rekowski P, Peletskaya E, et al. 1999. Experimental models of protein-RNA interaction: isolation and analyses of tRNA(Phe) and U1 snRNA-binding peptides from bacteriophage display libraries. J Neurosci Methods 18: 425–435 [DOI] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomito M, Wanner BL, Mori H 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2: 2006–0008. doi: 10.1038/msb4100050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björk G 1995. Biosynthesis and function of modified nucleosides. In tRNA: Structure, biosynthesis, and function (ed. Söll D and RajBhandary U), pp. 165–205 ASM Press, Washington [Google Scholar]
- Byrne R, Waterman D, Antson A 2009. Enzyme-RNA substrate recognition in RNA-modifying enzymes. In DNA and RNA modification enzymes: Structure, mechanisms, function, and evolution (ed. Grosjean Henri), pp. 304–327 Landes Bioscience, Austin [Google Scholar]
- Chen Y, Sierzputowska-Gracz H, Guenther R, Everett K, Agris PF 1993. 5-Methylcytidine is required for cooperative binding of Mg2+ and a conformational transition at the anticodon stem-loop of yeast phenylalanine tRNA. Biochemistry 32: 10249–10253 [DOI] [PubMed] [Google Scholar]
- Chen J, Ji C, Gu S, Zhao E, Dai J, Huang L, Qian J, Ying K, Xie Y, Mao Y 2003. Isolation and identification of a novel cDNA that encodes human yrdC protein. J Hum Genet 48: 164–169 [DOI] [PubMed] [Google Scholar]
- Chheda GB, Hong CI, Piskorz CF, Harmon GA 1972. Biosynthesis of N-(purin-6-ylcarbamoyl)-L-threonine riboside. Incorporation of L-threonine in vivo into modified nucleoside of transfer ribonucleic acid. Biochem J 127: 515–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chimnaronk S, Forouhar F, Sakai J, Yao M, Tron CM, Atta M, Fontecave M, Hunt JF, Tanaka I 2009. Snapshots of dynamics in synthesizing N6-isopentenyladenosine at the tRNA anticodon. Biochemistry 48: 5057–5065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A 1995. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biol NMR 6: 277–293 [DOI] [PubMed] [Google Scholar]
- Durant PC, Davis DR 1999. Stabilization of the anticodon stem-loop of tRNALys,3 by an A+-C base-pair and by pseudouridine. J Mol Biol 285: 115–131 [DOI] [PubMed] [Google Scholar]
- Durant PC, Bajji AC, Sundaram M, Kumar RK, Davis DR 2005. Structural effects of hypermodified nucleosides in the Escherichia coli and human tRNALys anticodon loop: the effect of nucleosides s2U, mcm5U, mcm5s2U, mnm5s2U, t6A, and ms2t6A. Biochemistry 44: 8078–8089 [DOI] [PubMed] [Google Scholar]
- Elkins BN, Keller EB 1974. The enzymatic synthesis of N-(Purin-6-ylcarbamoyl)threonine, an anticodon-adjacent base in transfer ribonucleic acid. Biochemistry 13: 4622–4628 [DOI] [PubMed] [Google Scholar]
- El Yacoubi B, Lyons B, Cruz Y, Reddy R, Nordin B, Agnelli F, Williamson JR, Schimmel P, Swairjo MA, de Crécy-Lagard V 2009. The universal Sua5/YciO/YrdC family is required for the formation of threonylcarbamoyladenosine in tRNA. Nucleic Acids Res 37: 2894–2909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia G, Goodenough-Lasua D 1998. Mechanisms of RNA-modifying and -editing enzymes. In Modification and editing of RNA (ed. Benne R and Grosjean H), pp. 135–168 ASM Press, Washington, DC [Google Scholar]
- Goddard TD, Kneller DG 2008. SPARKY 3, University of California, San Francisco [Google Scholar]
- Helm M 2006. Post-transcriptional nucleotide modification and alternate folding of tRNA. Nucleic Acids Res 34: 721–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitani R, Yokoyama S, Nureki O 2008. Structure, dynamics, and function of RNA modification enzymes. Curr Opin Struct Biol 18: 330–339 [DOI] [PubMed] [Google Scholar]
- Jühling F, Mörl M, Hartmann RK, Sprinzl M, Stadler PF, Pütz J 2009. tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res 37: D159–D162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaczanowska M, Rydén-Aulin M 2004. Temperature sensitivity caused by mutant release factor 1 is suppressed by mutations that affect 16S rRNA maturation. J Bacteriol 186: 3046–3055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaczanowska M, Rydén-Aulin M 2005. The YrdC protein—a putative ribosome maturation factor. Biochim Biophys Acta 1727: 87–96 [DOI] [PubMed] [Google Scholar]
- Kambampati R, Lauhon CT 2003. MnmA and IscS are required for in vitro 2-thiouridine biosynthesis in Escherichia coli. Biochemistry 42: 1109–1117 [DOI] [PubMed] [Google Scholar]
- Körner A, Söll D 1974. N-(Purin-6-ylcarbamoyl) threonine: Biosynthesis in vitro in transfer RNA by an enzyme purified from Escherichia coli. FEBS Lett 39: 301–306 [DOI] [PubMed] [Google Scholar]
- Kurganov B, Sugrobov N, Yakovlev V 1972. Estimation of dissociation constant of enzyme-ligand complex from fluorometric data by “difference” method. FEBS Lett 19: 308–310 [DOI] [PubMed] [Google Scholar]
- Lin CA, Ellis SR, True HL 2010. The Sua5 protein is essential for normal translational regulation in yeast. Mol Cell Biol 30: 354–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer M, Meyer B 1999. Characterization of ligand binding by saturation transfer difference NMR spectroscopy. Angew Chem Int Ed Engl 38: 1784–1788 [DOI] [PubMed] [Google Scholar]
- Miller JP, Hussain Z, Schweizer MP 1976. The involvement of the anticodon adjacent modified nucleoside N-[9-(β-D-ribofuranosyl) purine-6-ylcarbamoyl]-threonine in the biological function of E. coli tRNA-Ile. Nucleic Acids Res 3: 1185–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy FV, Ramakrishnan V, Malkiewicz A, Agris PF 2004. The role of modifications in codon discrimination by tRNALysUUU. Nat Struct Mol Biol 11: 1186–1191 [DOI] [PubMed] [Google Scholar]
- Na JG, Pinto I, Hampsey M 1992. Isolation and characterization of SUA5, a novel gene required for normal growth in Saccharomyces cerevisiae. Genetics 131: 791–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura S 1972. Minor components of transfer RNA: their characterization, localization, and function. Prog Nucleic Acid Res Mol Biol 12: 49–85 [PubMed] [Google Scholar]
- Numata T, Ikeuchi Y, Fukai S, Suzuki T, Nureki O 2006. Snapshots of tRNA sulphuration via an adenylated intermediate. Nature 442: 419–424 [DOI] [PubMed] [Google Scholar]
- Nureki O, Niimi T, Muramatsu T, Kanno H, Kohno T, Florentz C, Giegé R, Yokoyama S 1994. Molecular recognition of the identity-determinant set of isoleucine transfer RNA from Escherichia coli. J Mol Biol 236: 710–724 [DOI] [PubMed] [Google Scholar]
- Powers DM, Peterkofsky A 1972. Biosynthesis and specific labeling of N-(purin-6-ylcarbamoyl) threonine of Escherichia coli transfer RNA. Biochem Biophys Res Commun 46: 831–838 [DOI] [PubMed] [Google Scholar]
- Rachofsky EL, Osman R, Ross JB 2001. Probing structure and dynamics of DNA with 2-aminopurine: effects of local environment on fluorescence. Biochemistry 40: 946–956 [DOI] [PubMed] [Google Scholar]
- Roberts RJ, Cheng X 1998. Base flipping. Annu Rev Biochem 67: 181–198 [DOI] [PubMed] [Google Scholar]
- Scaringe SA, Wincott FE, Caruthers MH 1998. Novel RNA synthesis method using 5 ′-O-Silyl-2 ′-O-orthoester protecting groups. J Am Chem Soc 120: 11820–11821 [Google Scholar]
- Seif E, Hallberg BM 2009. RNA-protein mutually induced fit: structure of Escherichia coli isopentenyl-tRNA transferase in complex with tRNA(Phe). J Biol Chem 284: 6600–6604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan M, Mehta P, Yu Y, Prugar E, Koonin EV, Karzai AW, Sternglanz R 2011. The highly conserved KEOPS/EKC complex I essential for a universal tRNA modification, t6A. EMBO J 30: 873–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart JW, Gdaniec Z, Guenther R, Marszalek M, Sochacka E, Malkiewicz A, Agris PF 2000. Functional anticodon architecture of human tRNA Lys3 includes disruption of intraloop hydrogen bonding by the naturally occurring amino acid modification, t6A. Biochemistry 39: 13396–13404 [DOI] [PubMed] [Google Scholar]
- Sundaram M, Durant PC, Davis DR 2000. Hypermodified nucleosides in the anticodon of tRNALys stabilize a canonical U-turn structure. Biochemistry 39: 12575–12584 [DOI] [PubMed] [Google Scholar]
- Teplova M, Tereshko V, Sanishvili R, Joachimiak A, Bushueva T, Anderson WF, Egli M 2000. The structure of the yrdC gene product from Escherichia coli reveals a new fold and suggests a role in RNA binding. Protein Sci 9: 2557–2566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbonavicius J, Qian Q, Durand JM, Hagervall TG, Björk GR 2001. Improvement of reading frame maintenance is a common function for several tRNA modifications. EMBO J 20: 4863–4873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vendeix FAB, Graham WD, Dziergowska A, Gustilo E, Sproat B, Malkiewicz A, Agris PF 2008. Anticodon domain modifications contribute order to tRNA for ribosome-mediated codon binding. Biochemistry 47: 6117–6129 [DOI] [PubMed] [Google Scholar]
- Weissenbach J, Grosjean H 1981. Effect of threonylcarbamoyl modification (t6A) in yeast tRNA Arg III on codon-anticodon and anticodon-anticodon interactions. A thermodynamic and kinetic evaluation. Eur J Biochem 116: 207–213 [DOI] [PubMed] [Google Scholar]
- Yarian CS, Cain R, Basti MM, Ansari G, Guenther RH, Sochacka E, Malkiewicz A, Agris PF 1999. Structural and functional roles of the N1- and N3-protons of Ψ at tRNA's position 39. Nucleic Acids Res 27: 3543–3549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarian C, Townsend H, Czestkowski W, Sochacka E, Malkiewicz AJ, Guenther R, Miskiewicz A, Agris PF 2002. Accurate translation of the genetic code depends on tRNA modified nucleosides. J Biol Chem 277: 16391–16395 [DOI] [PubMed] [Google Scholar]