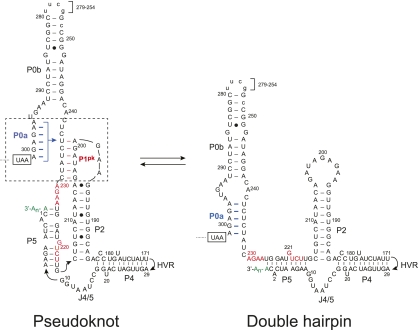
FIGURE 1.
Secondary structural representation of a proposed molecular switch model of the MHV 3′-UTR region consistent with functional studies (Goebel et al. 2004a; Zust et al. 2008). The psuedoknotted conformation is shown on the left, with the double hairpin conformation shown on the right. Formation of the P1pk helix (left, red base pairs) is mutually exclusive with the formation of the P0a stem (right, highlighted in blue). Nucleotides are numbered 3′ to 5′ from the 3′ terminus of the genome (C1) to A301, encompassing the entire 3′ UTR in MHV. Nucleotides that form the short P3 helix in the L1 loop (230AGAA227, 221GUCA218) (see Fig. 2A) are highlighted in red. The termination codon of the N gene is boxed, and in the RNAs investigated here, an extended stem–loop on P0b (nucleotides 279–254) not required for viral replication (Goebel et al. 2004a) was replaced with a uucg tetraloop sequence. The hypervariable region (HVR, nucleotides 170–30) is not shown and is also dispensable for viral replication in cells (Goebel et al. 2007). A 97-nt RNA that contains the minimal components of the proposed molecular switch corresponds to nucleotides 301–185 (Goebel et al. 2004a).