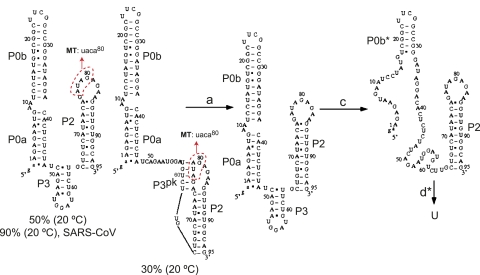
FIGURE 12.
The predicted structural changes associated with thermal unfolding of the 97-nt MHV UTR RNA calculated at 1 M NaCl. The two predominant conformations are shown with the percent of all conformations indicated at 20°C. The two conformers differ only in the disposition of nucleotides 78–82; in the major conformer these nucleotides pair to form the short stem–loop P3 contained entirely in loop L1. In the right conformer, these nucleotides pair with nucleotides 75–79 in the P2 loop to form a non-native pseudoknot, P3pk. In no case do nucleotides 41–49 of P0a pair with nucleotides in the P2 loop, as found for the PK RNA simulations (see Fig. 6). Simulations with the 92-nt SARS-CoV UTR RNA give rise to 90% of the conformations on the left, with no P1pk pseudoknot present. For the MHV UTR RNA, transition a is found to correspond to unfolding of the pseudoknot P3pk helix in the minor conformer, while b-transition unfolding is not present in either the simulated or experimental melts. Transition c corresponds to unfolding of the P0a stem and part of the P0b stem, and transition d is associated with unfolding of the P2 stem and the remainder of the P0b stem (denoted P0b*). The 4-nt substitution to create the UTR MT RNA is shown schematically on both major native-state conformers (circles in red).