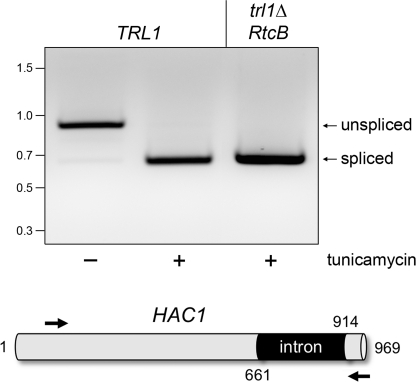
FIGURE 2.
RtcB can perform HAC1 mRNA splicing in the unfolded protein response. The unprocessed yeast HAC1 open reading frame, containing a 252-nucleotide intron, is depicted at the bottom. To gauge HAC1 mRNA splicing, cultures (10 ml) of S. cerevisiae trl1Δ p(2μ HIS3 TRL1) and trl1Δ p(2μ HIS3 RtcB) cells were grown at 30 °C in YPD media until the A600 reached 0.6. The cultures were split into 5-ml aliquots, one of which was adjusted to 1.5 μg/ml tunicamycin to induce endoplasmic reticulum stress. One hour later, the cells were harvested by centrifugation. Total RNA was isolated by using a yeast RNA purification kit (Epicentre Biotechnologies). HAC1 transcripts were detected by RT-PCR using primers flanking the intron. The cDNAs were synthesized by using the SuperScript III system (Invitrogen) with 1.0 μg of total RNA as template and 0.1 μm of HAC1-specific antisense primer (5′-dCATGAAGTGATGAAGAAATCATTCAATTC; complementary to nucleotides 940–968). The cDNAs were amplified by 22 cycles of PCR with Herculase (Stratagene) using the antisense primer and a sense-strand primer (5′-dCCAAGGAAAAGAGCCAAGACAAAAGAGG; corresponding to nucleotides 82–109). The RT-PCR products were analyzed by 1.4% agarose gel electrophoresis. A negative image of the ethidium bromide-stained gel is shown. The positions and sizes (in bp) of marker dsDNAs are indicated at left. The predicted sizes of the unspliced and spliced HAC1 RT-PCR products are 886 and 634 bp, respectively.