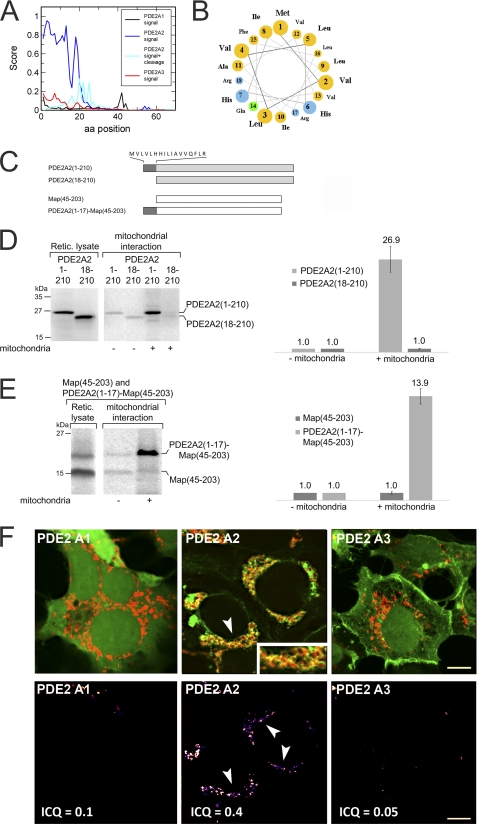
FIGURE 3.
The PDE2A2 N terminus acts as a mitochondrial localization signal. A, probability scores, calculated with SignalP, for the N termini of PDE2A isoforms 1 through 3 to act as mitochondrial localization sequences. A score combining this probability with the likelihood of a position to serve as a mitochondrial processing site (signal + cleavage) identifies three sites with a high probability to serve as proteolytic processing sites during import. aa, amino acid. B, a helical wheel representation of the 19 N-terminal residues of PDE2A2 reveals that these residues can form the amphipathic helix (top, hydrophobic; bottom, hydrophilic) typical for mitochondrial localization sequences. C, constructs used in the mitochondrial interaction experiments shown in panels D and E. Protein parts from the mitochondrial matrix protein Map are symbolized as white bars, residues from human PDE2A2 are symbolized as gray bars, and the 18 N-terminal residues unique for this PDE2A isoform (sequence on top) are symbolized as dark gray bars. D, in vitro tests for interaction with mitochondria. Radiolabeled samples (left) of PDE2A2(1–210) and PDE2A2(18–210) were incubated with rat liver mitochondria for 10 min at 25 °C. Mitochondria were reisolated, and associated proteins were analyzed by SDS-PAGE and autoradiography using a BAS-1800 II imager (left). For quantitation (right), the amount of protein in the absence of rat liver mitochondria was set to a value of 1 (control). Retic. lysate, reticulocyte lysate. E, in vitro tests for the interaction of Map(45–203) and PDE2A2(1–17)-Map(45–203) with mitochondria. The experiments were carried out as described in panel D. Error bars in panels D and E represent S.D. values. F, upper panel, confocal images of HEK cells overexpressing a mitochondrial marker (DsRed-mito, red) and the N termini of PDE2 isoforms fused to EGFP or CFP (green). The three PDE2A isoforms show distinct subcellular distributions, and considerable overlap with the mitochondrial marker, visible in yellow (see for example the arrow), is specifically observed for isoform 2 (see inset). Lower panel, graphical presentation of the intensity correlation analysis highlighting substantial overlap of the PDE2A2 isoform with DsRed-mito. Scale bar: 3 μm.