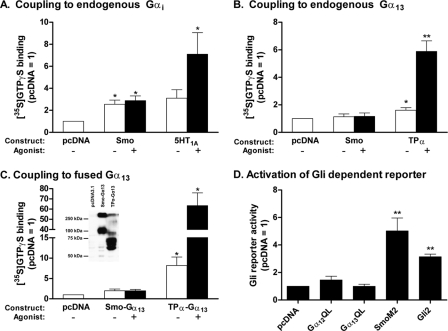
FIGURE 1.
Smoothened does not couple to Gα13 and does not activate Gli(s) in HEK293 cells. A, HEK293 cells were transfected with vector for Smo or the 5-HT1A receptor or with empty vector (pcDNA). At 48 h, membranes were isolated and [35S]GTPγS binding to endogenous Gαi was evaluated with or without 10 μm purmorphamine (Smo) or 1 μm 8-OH-DPAT (5-HT1A receptor). [35S]GTPγS binding is normalized to levels obtained with empty vector and represents four to five individual experiments performed in triplicate. B, HEK293 cells were transfected with vector for Smo or TPα, or with empty vector, and [35S]GTPγS binding to endogenous Gα13 was evaluated as above with or without 10 μm purmorphamine (Smo) or U46619 (TPα). The data represent four individual experiments performed in triplicate. C, HEK293 cells were transfected with vector for Smo-Gα13 or TPα-Gα13, or with empty vector, and [35S]GTPγS binding was evaluated with or without 10 μm purmorphamine (Smo) or U46619 (TPα). The data represent four individual experiments performed in triplicate. Inset, Western blot showing the relative expression levels of receptor·Gα fusion proteins present in membranes using a Gα13-directed antibody. Molecular weights inferred from cDNA for Smo-Gα13 and TPα-Gα13 are 103,100 and 82,700, respectively; heterogeneity in banding is presumably due to oligomerization, varying degrees of glycosylation, and/or multiple initiation sites. D, HEK293 cells were transfected with vector for constitutively active Gα12 or Gα13 (QL mutants), for SmoM2 or Gli2, or with empty vector (pcDNA), together with 8xGli firefly and TK Renilla luciferase reporters. At 24 h, when cells reached confluence, the serum was lowered to 0.5%. Luciferase activities were assayed 24 h later. The data are means of ratios of firefly/Renilla activities normalized to those obtained for pcDNA from six independent experiments carried out in triplicate. For all panels in this figure, significance was determined using paired t tests. *, p < 0.05 and **, p < 0.01.