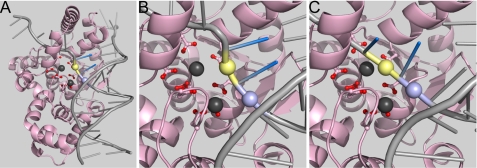
FIGURE 5.
Proposed double nucleotide unpairing in T5FEN catalyzed reactions. A, model of T5FEN (1UT5) bound to a pseudo-Y DNA created by overlay with the structure of T4 RNase H (2IHN, T4FEN) bound to DNA (T4 protein not shown). Active site carboxylates are shown as sticks with red oxygens, and T5 metal ions (M2 foreground) are shown in gray. The duplex where the reaction occurs is bound in front of and parallel to the active site. The protein-DNA structure 2IHN was solved in the absence of metal ions; hence the substrate arches away and does not occupy the carboxylate-rich active site. The scissile phosphate diester bond positioned 7 Å from M1 is colored yellow, and the adjacent duplex phosphate −1 is colored lilac. The two 5′-terminal nucleobases of the duplex are depicted in blue. B, close up of the active site in this model with coloring as in A. C, proposed unpairing of the two terminal base pairs of the duplex to allow the scissile phosphate diester (yellow) to move toward M1. This relocation requires that the adjacent phosphate diester −1 moves farther toward the carboxylate active site and closer to M2. In other FENs and related superfamily members, the N terminus plays a similar role to M2 (2, 4).